|
Dalton (unit)
The dalton or unified atomic mass unit (symbols: Da or u) is a unit of mass widely used in physics and chemistry. It is defined as 1/12 of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. The atomic mass constant, denoted mu is defined identically, giving mu = m(12C)/12 = 1 Da.
This unit is commonly used in physics and chemistry to express the mass of atomic-scale objects, such as atoms, molecules, and elementary particles, both for discrete instances and multiple types of ensemble averages. For example, an atom of helium-4 has a mass of 4.0026 Da. This is an intrinsic property of the isotope and all helium-4 have the same mass. Acetylsalicylic acid (aspirin), C9H8O4, has an average mass of approximately 180.157 Da. However, there are no acetylsalicylic acid molecules with this mass. The two most common masses of individual acetylsalicylic acid molecules are 180.04228 Da and 181.04565 Da.
The molecular masses of proteins, nucleic acids, and other large polymers are often expressed with the units kilodaltons (kDa), megadaltons (MDa), etc. Titin, one of the largest known proteins, has a molecular mass of between 3 and 3.7 megadaltons. The DNA of chromosome 1 in the human genome has about 249 million base pairs, each with an average mass of about 650 Da, or 156 GDa total. (W) |
|
degradosome
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.
The store of cellular RNA in the cells is constantly fluctuating. For example, in Escherichia coli, Messenger RNA's life expectancy is between 2 and 25 minutes, in other bacteria it might last longer. Even in resting cells, RNA is degraded in a steady state, and the nucleotide products of this process are later reused for fresh rounds of nucleic acid synthesis. RNA turnover is very important for gene regulation and quality control.
All organisms have various tools for RNA degradation, for instance ribonucleases, helicases, 3'-end nucleotidyltransferases (which add tails to transcripts), 5'-end capping and decapping enzymes and assorted RNA-binding proteins that help to model RNA for presentation as substrate or for recognition. Frequently, these proteins associate into stable complexes in which their activities are coordinate or cooperative. Many of these RNA metabolism proteins are represented in the components of the multi-enzyme RNA degradosome of Escherichia coli, which is constituted by four basic components: the hydrolytic endo-ribonuclease RNase E, the phosphorolytic exo-ribonuclease PNPase, the ATP-dependent RNA helicase (RhIB) and a glycolytic enzyme enolase.
The RNA degradosome was discovered in two different laboratories while they were working on the purification and characterization of E. coli, RNase E and the factors that could have an influence on the activity of the RNA-degrading enzymes, concretely, PNPase. It was found while two of its major compounds were being studied. (W)
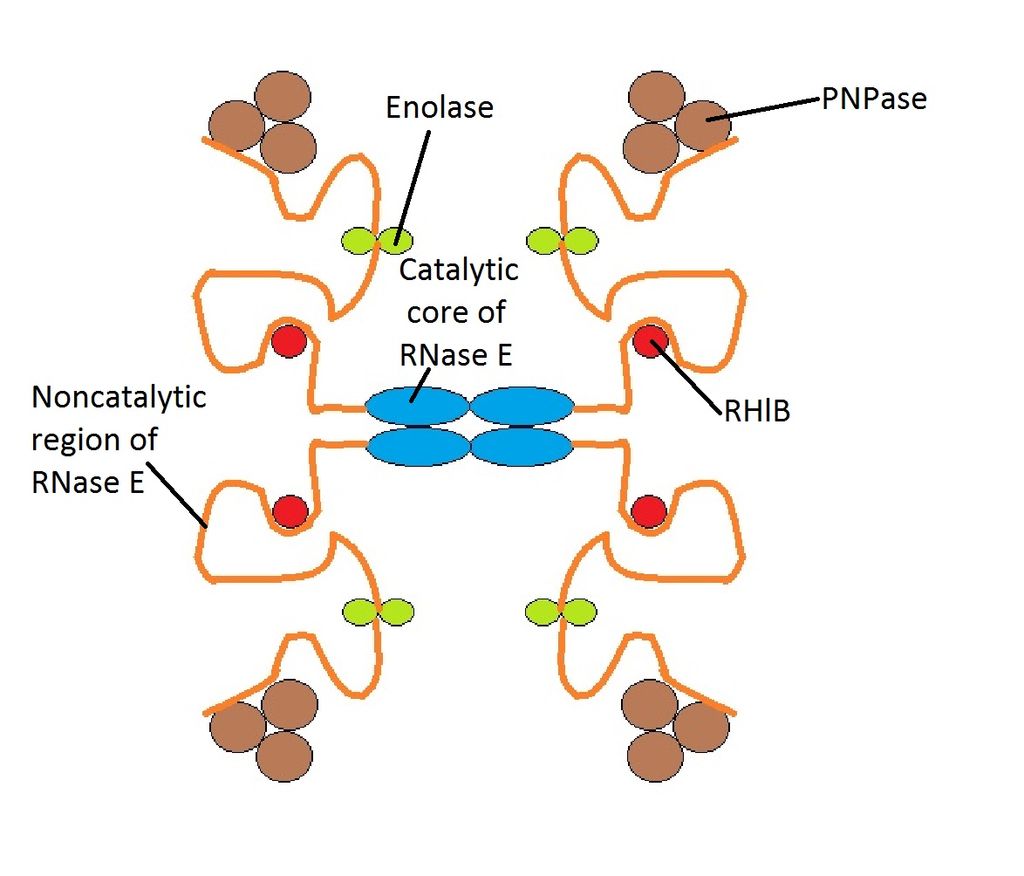
This would represent the basic structure of RNA Degradosome. The structure has been drawn symmetrically, however, it's a dynamic structure so the noncatalytic region of RNase E would form a random coil, and each of these coils would act independently from the other ones. |
|
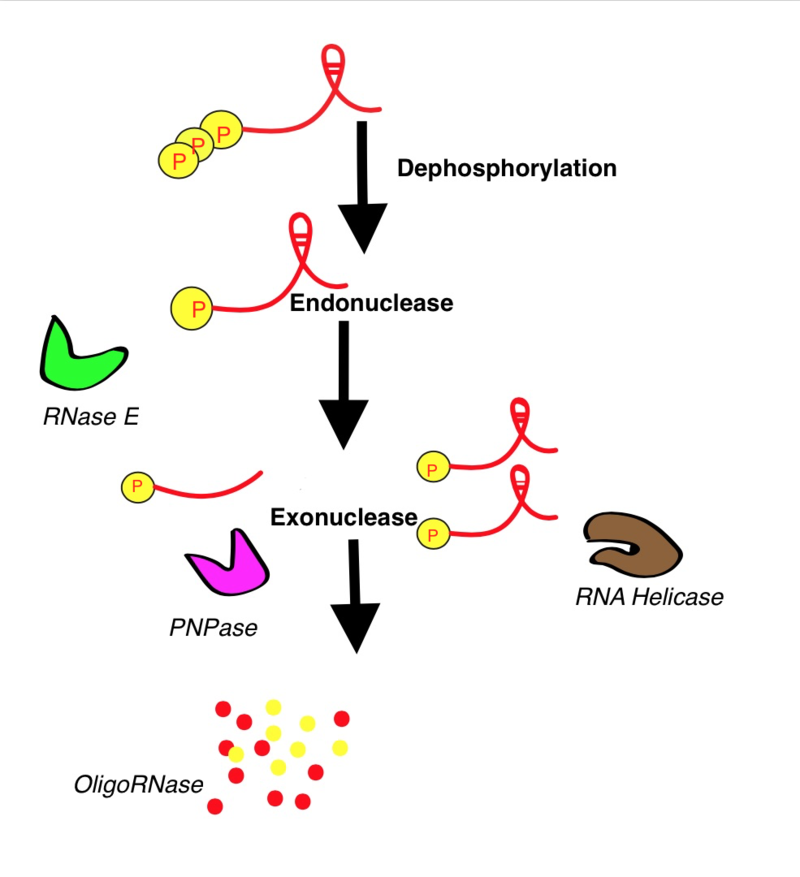
This picture shows RNA's degradation process with the specific phases. |
|
|
|
deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome.
The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slippage, within the DNA polymerase active site.
Deletions can be caused by errors in chromosomal crossover during meiosis, which causes several serious genetic diseases. Deletions that do not occur in multiples of three bases can cause a frameshift by changing the 3-nucleotide protein reading frame of the genetic sequence. Deletions are representative of eukaryotic organisms, including humans and not in prokaryotic organisms, such as bacteria. (W)
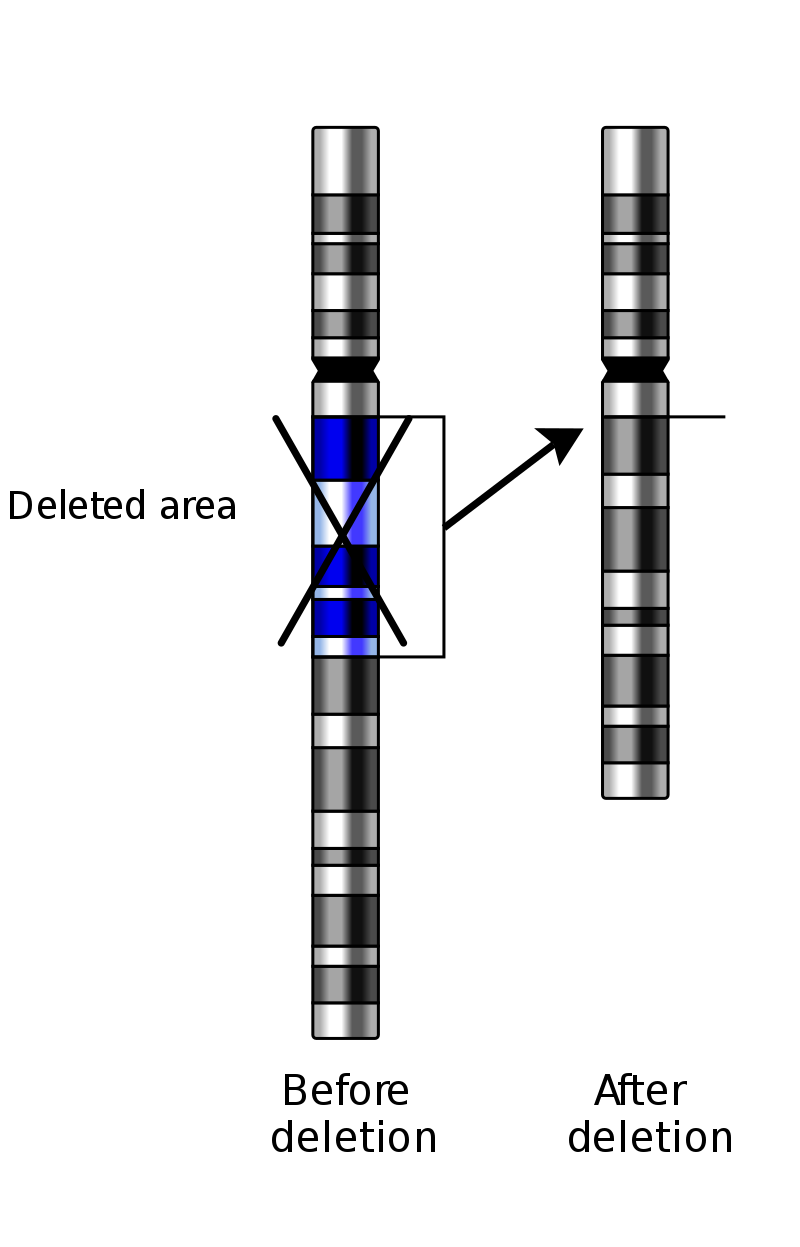
Deletion on a chromosome. |
|
|
|
denaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent (e.g., alcohol or chloroform), radiation or heat.If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups. Denatured proteins lose their 3D structure and therefore cannot function.
Protein folding is key to whether a globular or membrane protein can do its job correctly; it must be folded into the right shape to function. However, hydrogen bonds, which play a big part in folding, are rather weak and thus easily affected by heat, acidity, varying salt concentrations, and other stressors which can denature the protein. This is one reason why homeostasis is physiologically necessary in many life forms.
This concept is unrelated to denatured alcohol, which is alcohol that has been mixed with additives to make it unsuitable for human consumption. (W)

Spiegelei – Das Protein (Eiweiß) erfährt durch Zufuhr von Energie in Form von Wärme (Braten) eine Denaturierung (Gerinnung). |
|
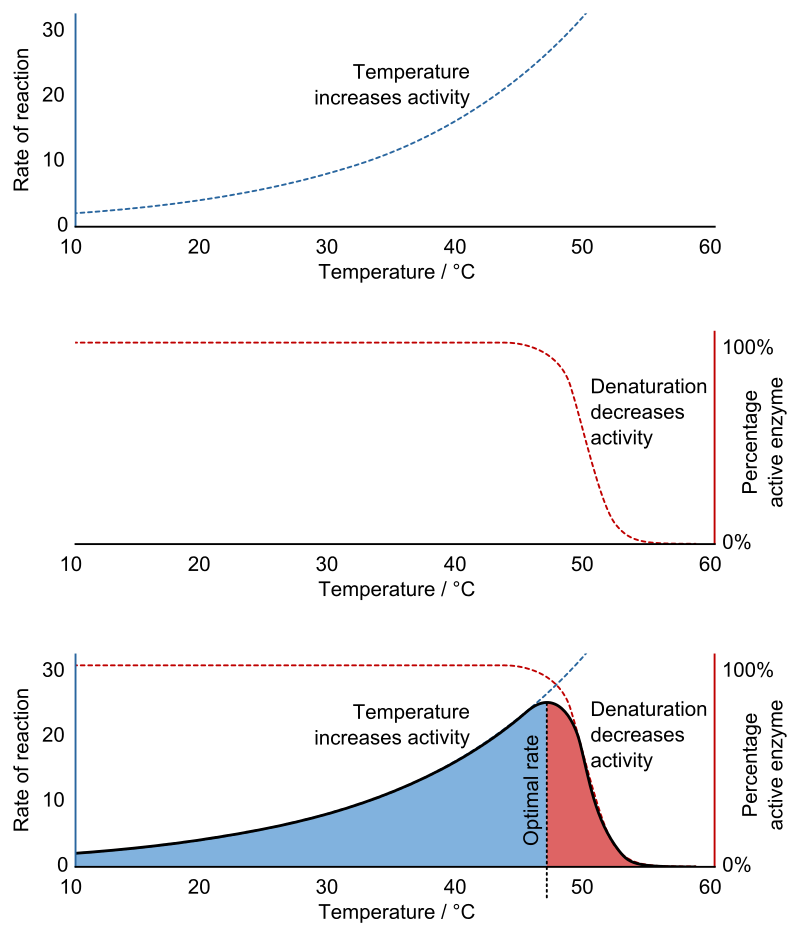
The effects of temperature on enzyme activity. Top - increasing temperature increases the rate of reaction (Q10 coefficient). Middle - the fraction of folded and functional enzyme decreases above its denaturation temperature. Bottom - consequently, an enzyme's optimal rate of reaction is at an intermediate temperature. |
|
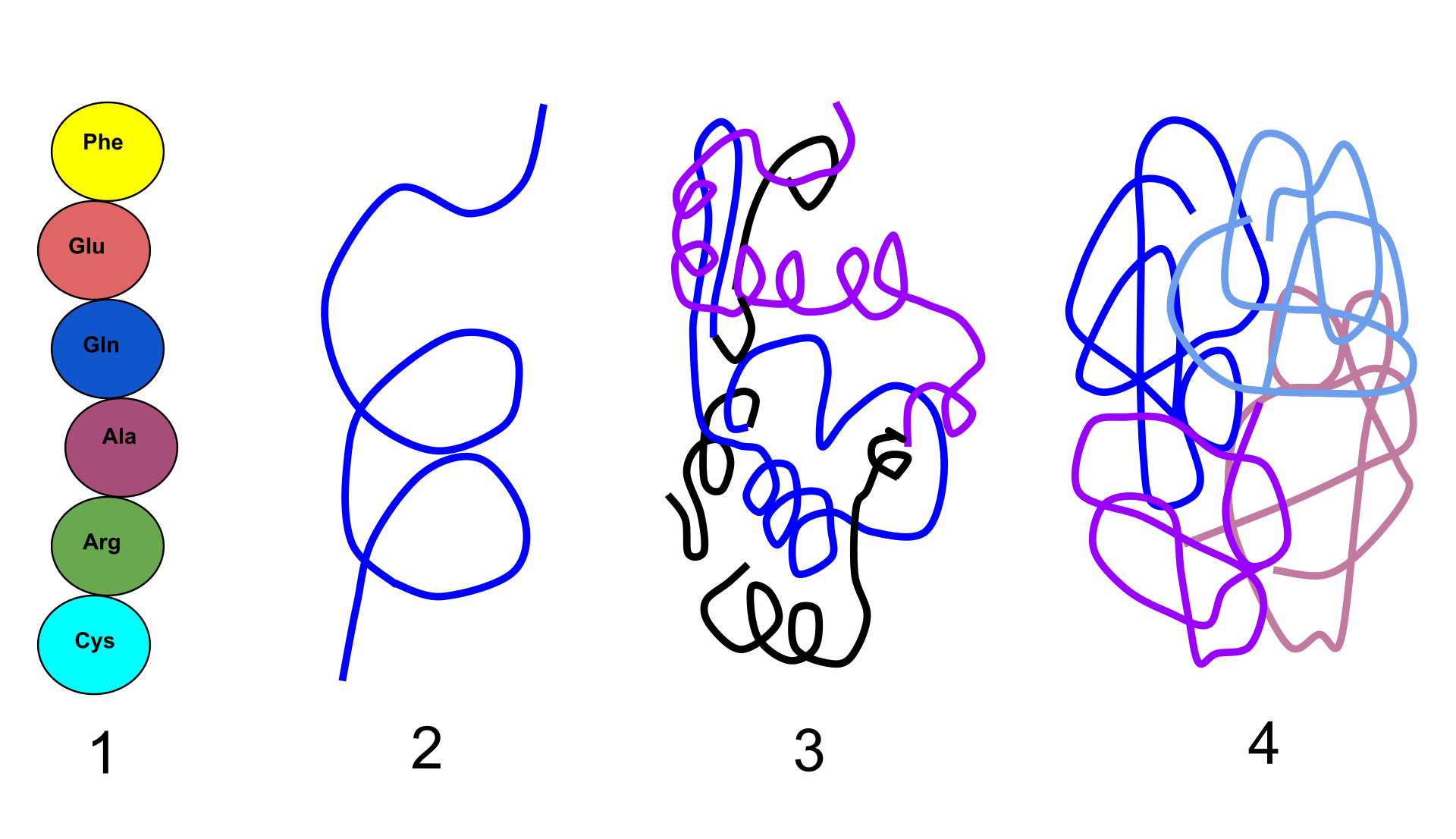
Functional proteins have four levels of structural organization: 1) Primary structure: the linear structure of amino acids in the polypeptide chain 2) Secondary structure: hydrogen bonds between peptide group chains in an alpha helix or beta sheet 3) Tertiary structure: three-dimensional structure of alpha helixes and beta helixes folded 4) Quaternary structure: three-dimensional structure of multiple polypeptides and how they fit together. |
|

(Top) The protein albumin in the egg white undergoes denaturation and loss of solubility when the egg is cooked. (Bottom) Paperclips provide a visual analogy to help with the conceptualization of the denaturation process. |
|
|
|
deoxyribonuclease
A deoxyribonuclease (DNase, for short) is an enzyme that catalyzes the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. Deoxyribonucleases are one type of nuclease, a generic term for enzymes capable of hydrolyzing phosphodiester bonds that link nucleotides. A wide variety of deoxyribonucleases are known, which differ in their substrate specificities, chemical mechanisms, and biological functions. (W)
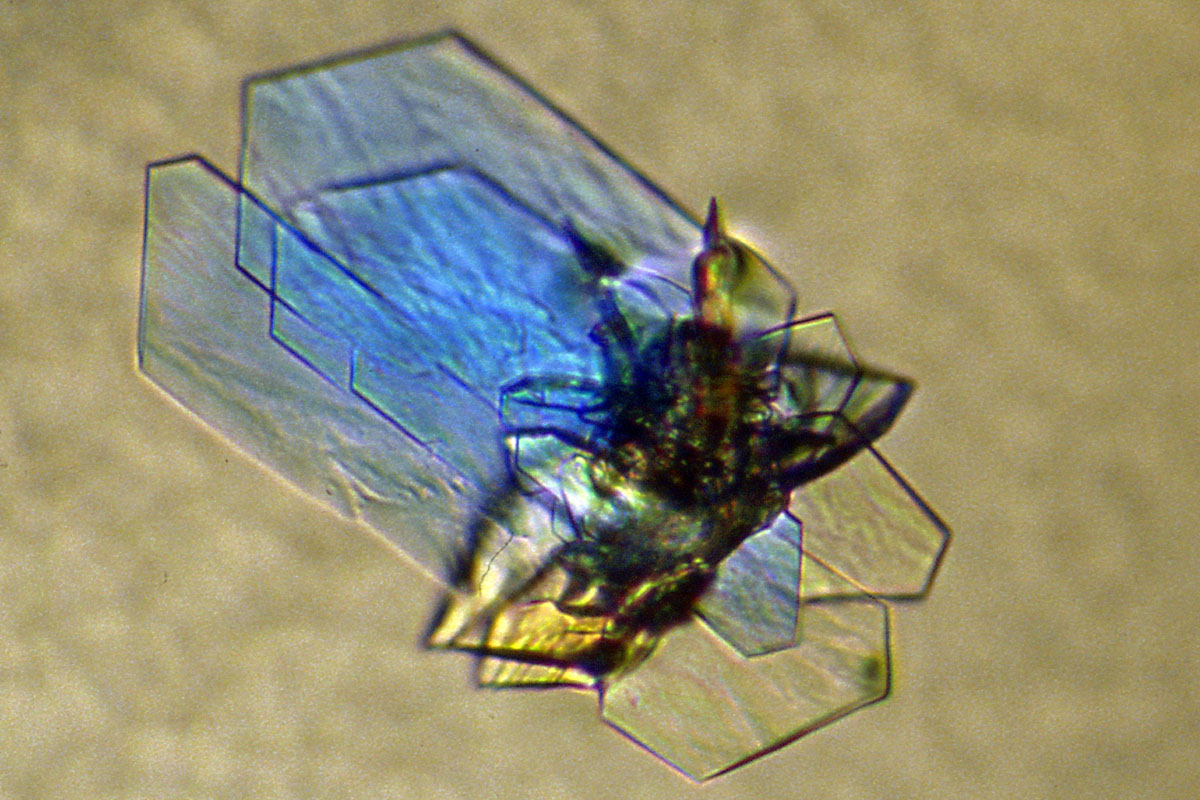
Crystals of a DNase protein. |
|
|
|
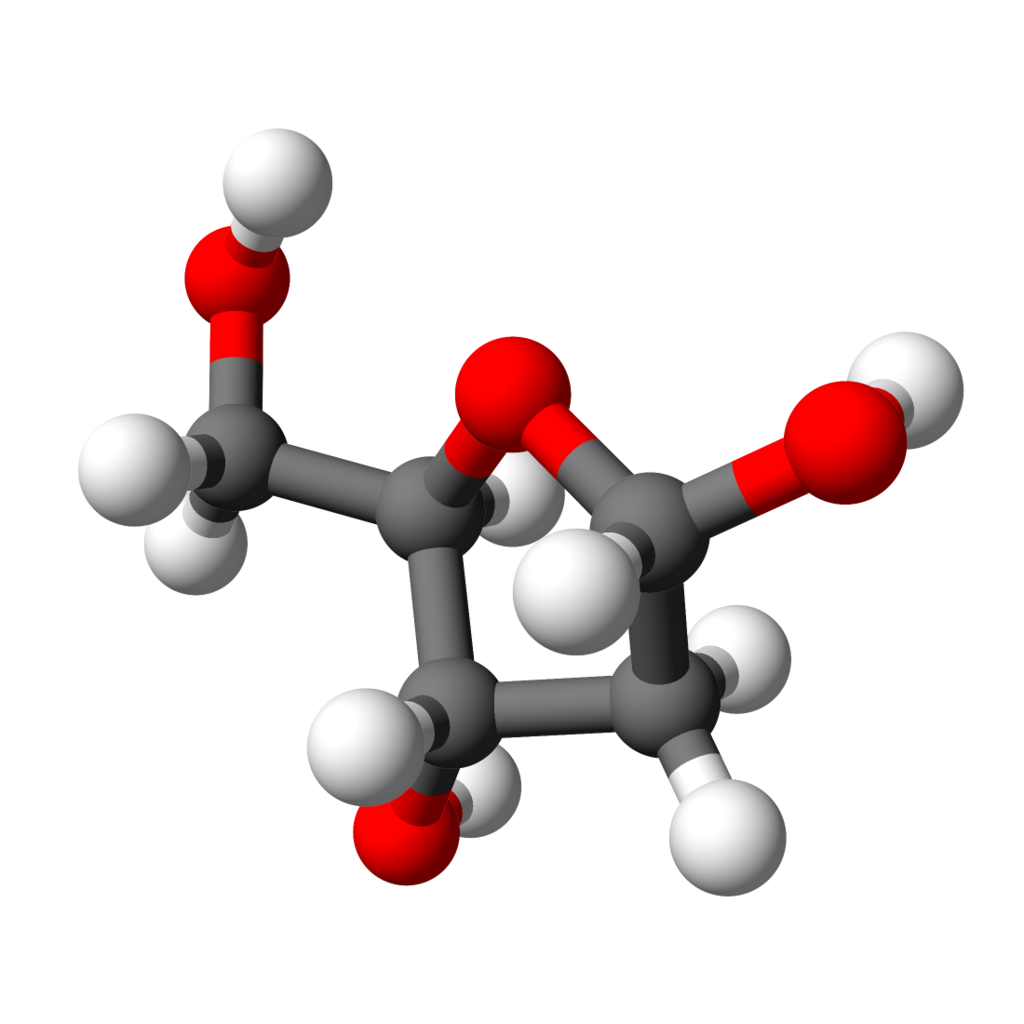
deoxyribose C5H10O4
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of an oxygen atom. (W)
🛑 deoxyribose
- Monosakkarid.
- DNA’nın ön-bileşeni.
- Bir DNA nükleotidi = deoxyriboz + organik baz (1′ riboz karbonuna bağlı adenin (A), thymin (T), guanin (G) ya da cytosine (C)).
|
|
|
|
|
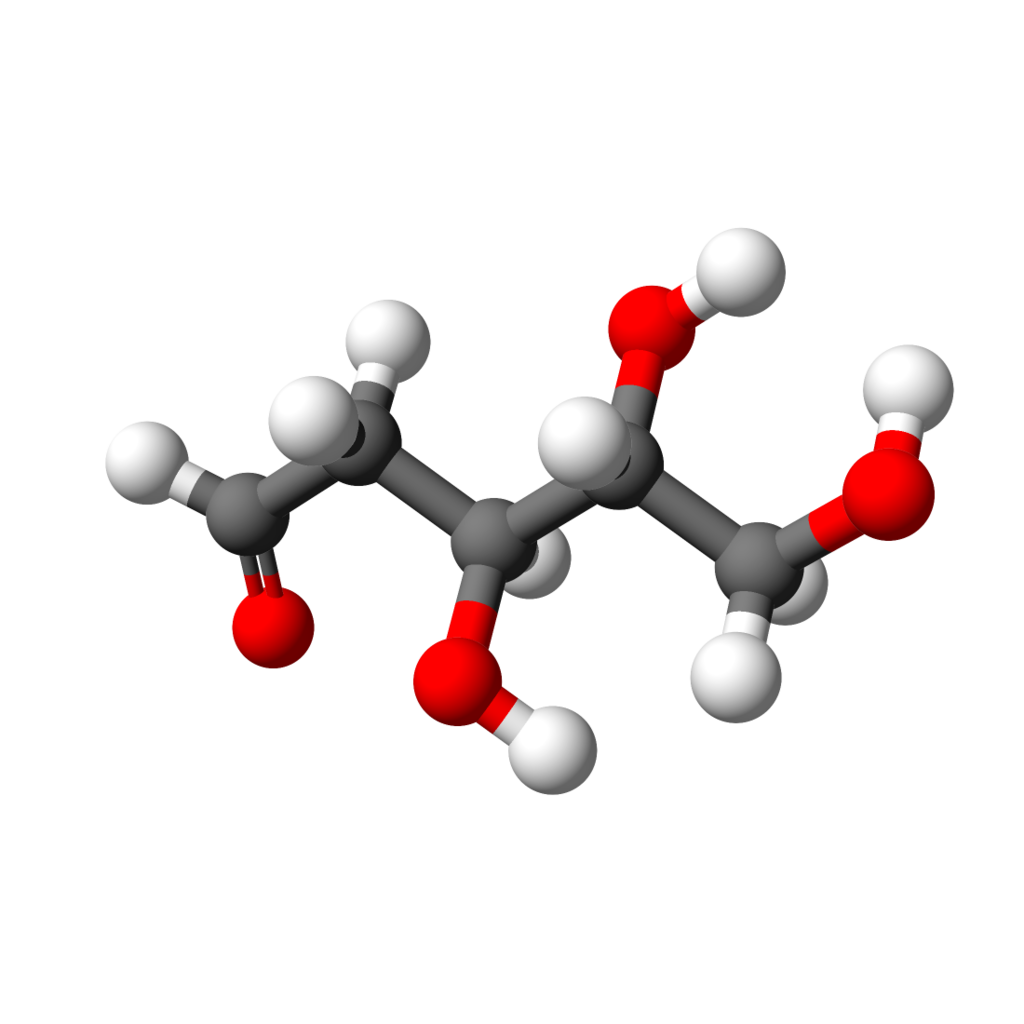
A model of D-deoxyribose in chain form. |
|
|
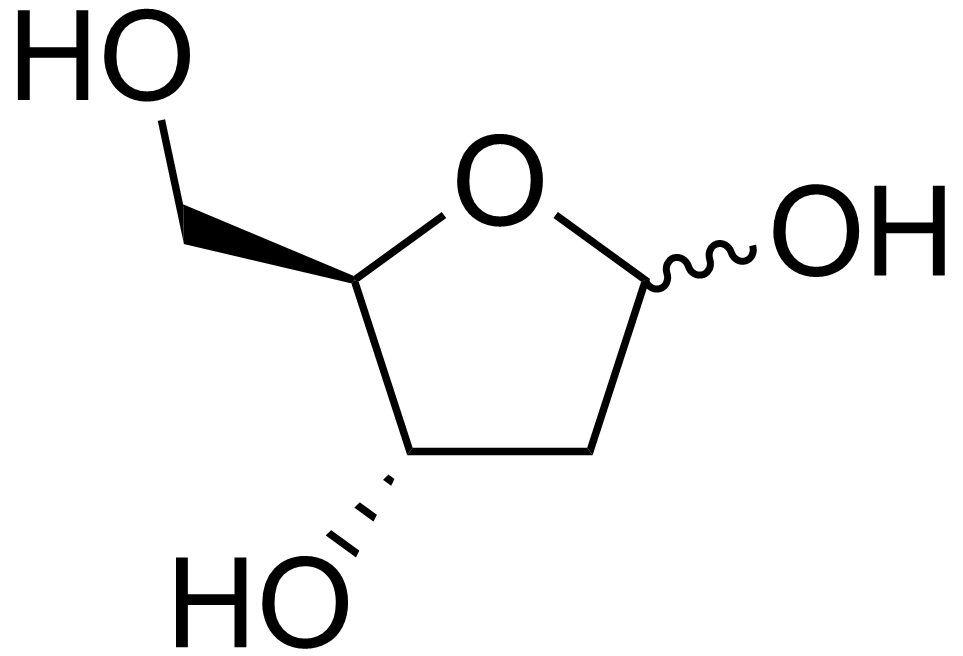
Chemical structure of D-deoxyribose. |
|
A model of D-deoxyribose. |
|
|
| |
📂 Deoxyribose
Deoxyribose C5H10O4 (W)
| Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of an oxygen atom. Since the pentose sugars arabinose and ribose only differ by the stereochemistry at C2′, 2-deoxyribose and 2-deoxyarabinose are equivalent, although the latter term is rarely used because ribose, not arabinose, is the precursor to deoxyribose. |
History
Deoxyribose was discovered in 1929 by Phoebus Levene.
Structure
Several isomers exist with the formula H−(C=O)−(CH2)−(CHOH)3−H, but in deoxyribose all the hydroxyl groups are on the same side in the Fischer projection. The term "2-deoxyribose" may refer to either of two enantiomers: the biologically important d-2-deoxyribose and to the rarely encountered mirror image l-2-deoxyribose. d-2-deoxyribose is a precursor to the nucleic acid DNA.2-deoxyribose is an aldopentose, that is, a monosaccharide with five carbon atoms and having an aldehyde functional group.
In aqueous solution, deoxyribose primarily exists as a mixture of three structures: the linear form H−(C=O)−(CH2)−(CHOH)3−H and two ring forms, deoxyribofuranose ("C3′-endo"), with a five-membered ring, and deoxyribopyranose ("C2′-endo"), with a six-membered ring. The latter form is predominant (whereas the C3′-endo form is favored for ribose).
.PNG)
Chemical equilibrium of deoxyribose in solution |
|
Biological importance
As a component of DNA, 2-deoxyribose derivatives have an important role in biology. The DNA (deoxyribonucleic acid) molecule, which is the main repository of genetic information in life, consists of a long chain of deoxyribose-containing units called nucleotides, linked via phosphate groups. In the standard nucleic acid nomenclature, a DNA nucleotide consists of a deoxyribose molecule with an organic base (usually adenine, thymine, guanine or cytosine) attached to the 1′ ribose carbon. The 5′ hydroxyl of each deoxyribose unit is replaced by a phosphate (forming a nucleotide) that is attached to the 3′ carbon of the deoxyribose in the preceding unit.
The absence of the 2′ hydroxyl group in deoxyribose is apparently responsible for the increased mechanical flexibility of DNA compared to RNA, which allows it to assume the double-helix conformation, and also (in the eukaryotes) to be compactly coiled within the small cell nucleus. The double-stranded DNA molecules are also typically much longer than RNA molecules. The backbone of RNA and DNA are structurally similar, but RNA is single stranded, and made from ribose as opposed to deoxyribose.
Other biologically important derivatives of deoxyribose include mono-, di-, and triphosphates, as well as 3′-5′ cyclic monophosphates.
Biosynthesis
Deoxyribose is generated from ribose 5-phosphate by enzymes called ribonucleotide reductases. These enzymes catalyse the deoxygenation process. |
|

|
|
|
|
|
dephosphorylation
Dephosphorylation is the removal of a phosphate (PO43-) group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and deactivate enzymes by detaching or attaching phosphoric esters and anhydrides. A notable occurrence of dephosphorylation is the conversion of ATP to ADP and inorganic phosphate.
Dephosphorylation employs a type of hydrolytic enzyme, or hydrolase, which cleave ester bonds. The prominent hydrolase subclass used in dephosphorylation is phosphatase. Phosphatase removes phosphate groups by hydrolysing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl (-OH) group.
The reversible phosphorylation-dephosphorylation reaction occurs in every physiological process, making proper function of protein phosphatases necessary for organism viability. Because protein dephosphorylation is a key process involved in cell signalling, protein phosphatases are implicated in conditions such as cardiac disease, diabetes, and Alzheimer's disease. (W)
|
|
dipeptide
A dipeptide is an organic compound derived from two amino acids. The constituent amino acids can be the same or different. When different, two isomers of the dipeptide are possible, depending on the sequence. Several dipeptides are physiologically important, and some are both physiologically and commercially significant. A well known dipeptide is aspartame, an artificial sweetener. (W)
|
|
direct DNA damage
Direct DNA damage can occur when DNA directly absorbs a UVB photon, or for numerous other reasons. UVB light causes thymine base pairs next to each other in genetic sequences to bond together into pyrimidine dimers, a disruption in the strand, which reproductive enzymes cannot copy. It causes sunburn and it triggers the production of melanin.
Other names for the "direct DNA damage" are:
Due to the excellent photochemical properties of DNA, this nature-made molecule is damaged by only a tiny fraction of the absorbed photons. DNA transforms more than 99.9% of the photons into harmless heat (but the damage from the remaining < 0.1% is still enough to cause sunburn). The transformation of excitation energy into harmless heat occurs via a photochemical process called internal conversion. In DNA, this internal conversion is extremely fast, and therefore efficient. This ultrafast (subpicosecond) internal conversion is a powerful photoprotection provided by single nucleotides. However, the Ground-State Recovery is much slower (picoseconds) in G·C−DNA duplexes and hairpins. It is presumed to be even slower for double-stranded DNA in conditions of the nucleus. The absorption spectrum of DNA shows a strong absorption for UVB radiation and a much lower absorption for UVA radiation. Since the action spectrum of sunburn is indistinguishable from the absorption spectrum of DNA, it is generally accepted that the direct DNA damages are the cause of sunburn. While the human body reacts to direct DNA damages with a painful warning signal, no such warning signal is generated from indirect DNA damage. (W)
|
|
directionality (molecular biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide sugar-ring means that there will be a 5′-end (usually pronounced "five prime end" ), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′-end (usually pronounced "three prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. (W)
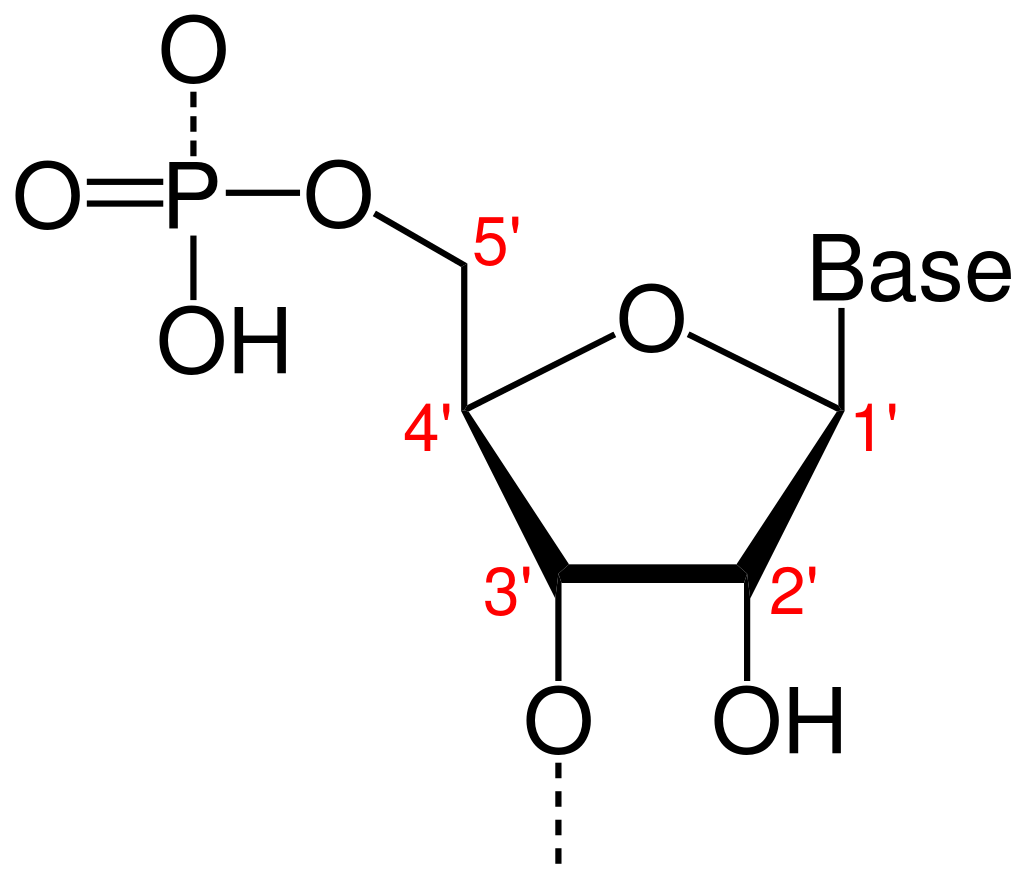
A furanose (sugar-ring) molecule with carbon atoms labeled using standard notation. The 5′ is upstream; the 3′ is downstream. DNA and RNA are synthesized in the 5′ to 3′ direction.. |
|
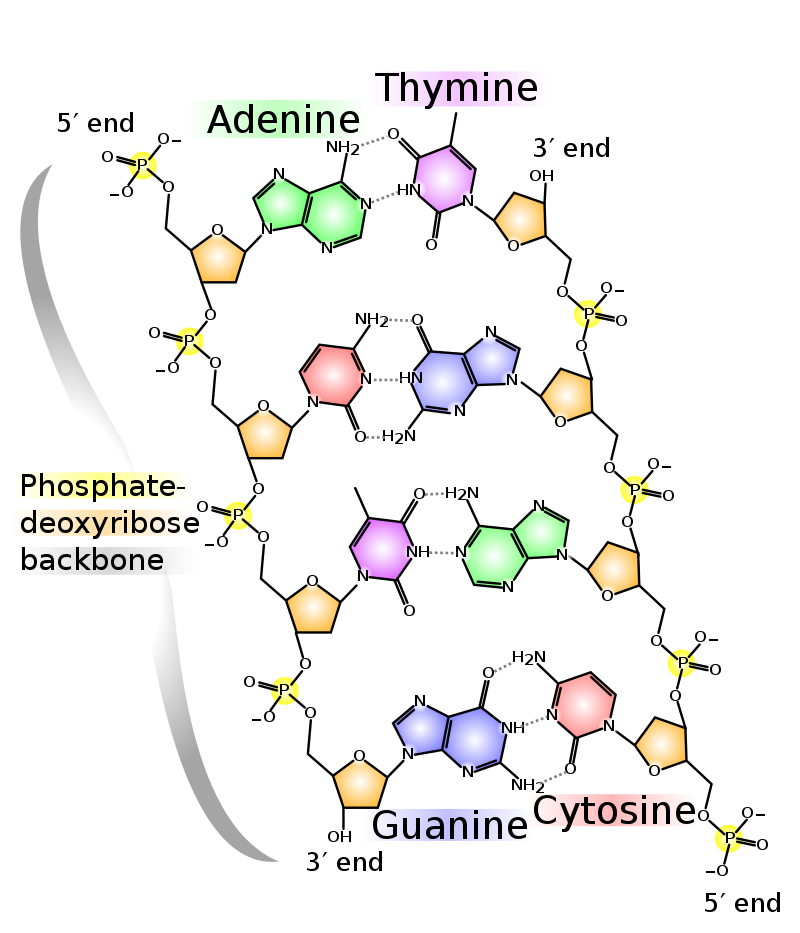
In the DNA segment shown, the 5′ to 3′ directions are down the left strand and up the right strand. |
|
|
|
disaccharide
A disaccharide (also called a double sugar or bivose) is the sugar formed when two monosaccharides (simple sugars) are joined by glycosidic linkage. Like monosaccharides, disaccharides are soluble in water. Three common examples are sucrose, lactose, and maltose. (W)
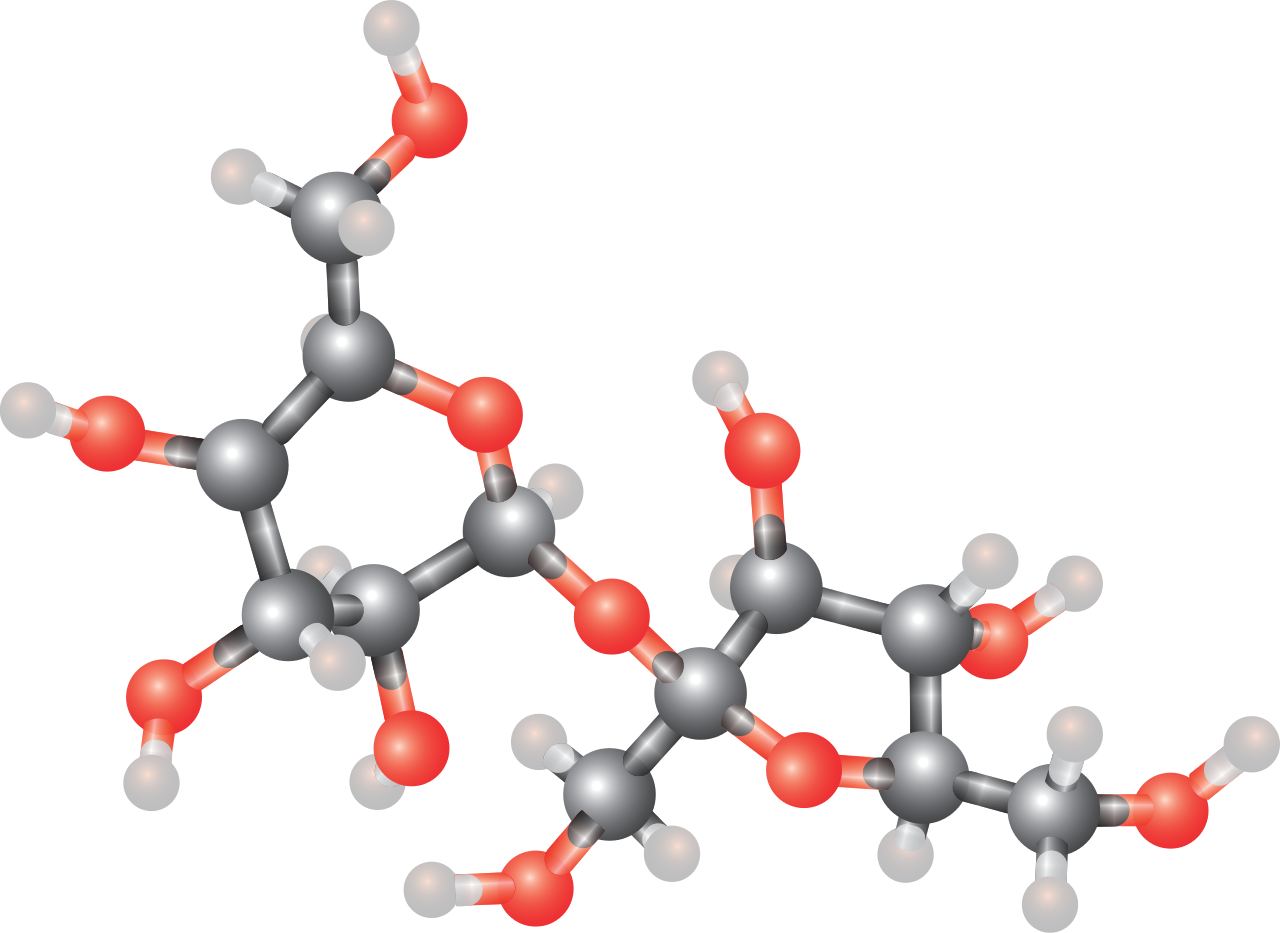
Sucrose, a disaccharide formed from condensation of a molecule of glucose and a molecule of fructose. |
|
|
|
disulfides, organic
Symmetrical disulfides are compounds of the formula R2S2. Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula RSSR'. They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. (W)
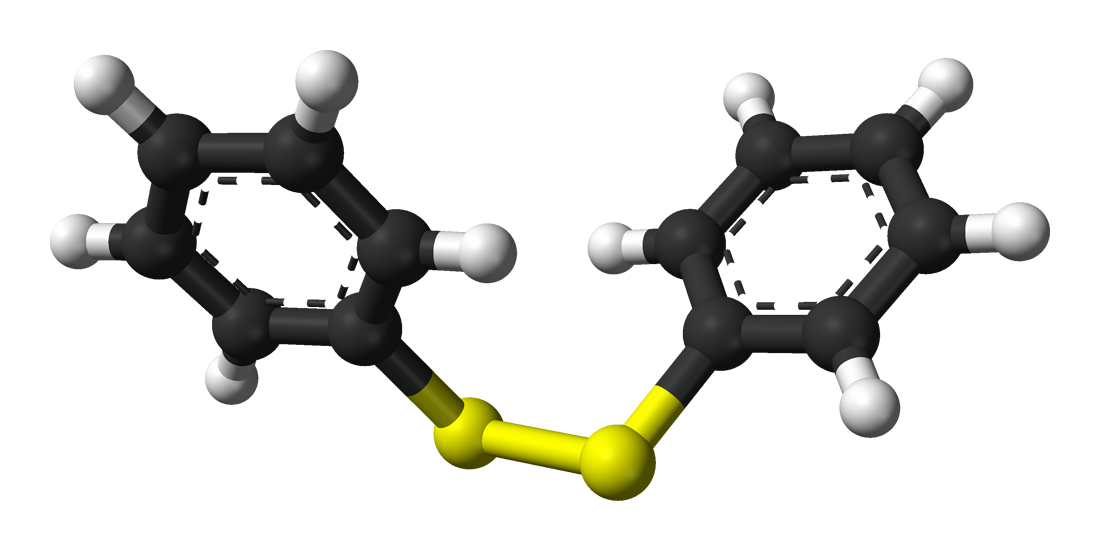
Ph2S2, a common organic disulfide. |
|
|
|
|
Deoxyribonucleic acid (DNA) is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life. (W)
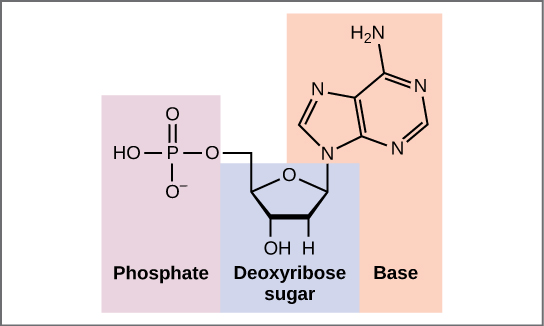
(a) Each DNA nucleotide is made up of a sugar, a phosphate group, and a base. (b) Cytosine and thymine are pyrimidines. Guanine and adenine are purines. (L) |
|
 forms a double stranded helix, and (b) adenine pairs with thymine and cytosine pairs with guanine.jpg)
DNA (a) forms a double stranded helix, and (b) adenine pairs with thymine and cytosine pairs with guanine. (L) |
|
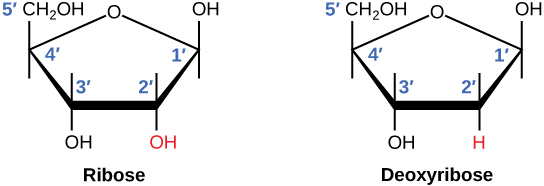
The difference between the ribose found in RNA and the deoxyribose found in DNA is that ribose has a hydroxyl group at the 2' carbon. (L) |
|
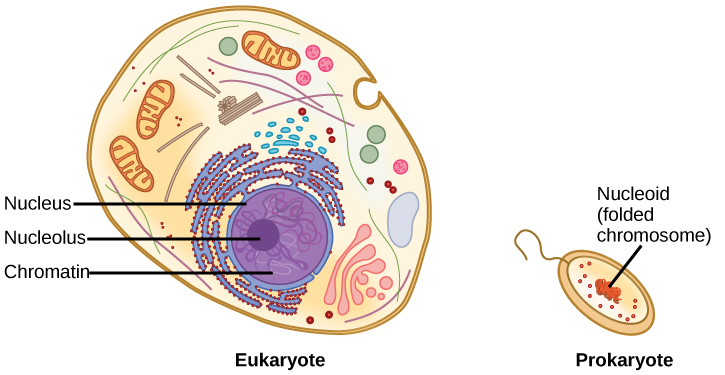
A eukaryote contains a well-defined nucleus, whereas in prokaryotes, the chromosome lies in the cytoplasm in an area called the nucleoid. (L) |
|
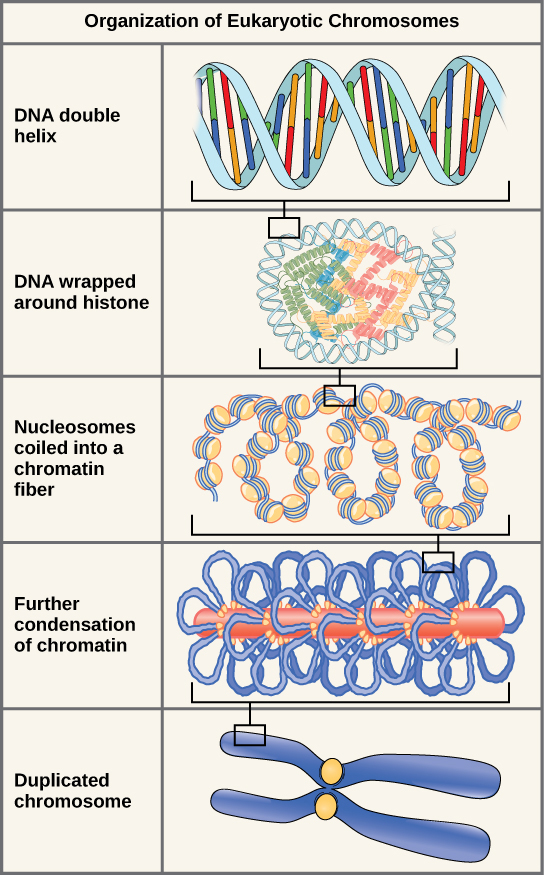
These figures illustrate the compaction of the eukaryotic chromosome. (L)
Each chromosome consists of one continuous thread-like molecule of DNA coiled tightly around proteins, and contains a portion of the 6,400,000,000 basepairs (DNA building blocks) that make up your DNA. (L) |
|
|
|
DNA adduct
In molecular genetics, a DNA adduct is a segment of DNA bound to a cancer-causing chemical. This process could be the start of a cancerous cell, or carcinogenesis. DNA adducts in scientific experiments are used as biomarkers of exposure and as such are themselves measured to reflect quantitatively, for comparison, the amount of carcinogen exposure to the subject organism, for example rats or other living animals. Under experimental conditions for study, such DNA adducts are induced by known carcinogens, of which commonly used is DMBA (7,12-dimethylbenz(a)anthracene). For example, the term "DMBA-DNA adduct" in a scientific journal refers to a piece of DNA that has DMBA attached to it. The presence of such an adduct indicates prior exposure to a potential carcinogen, but does not by itself indicate the presence of cancer in the subject animal. (W)
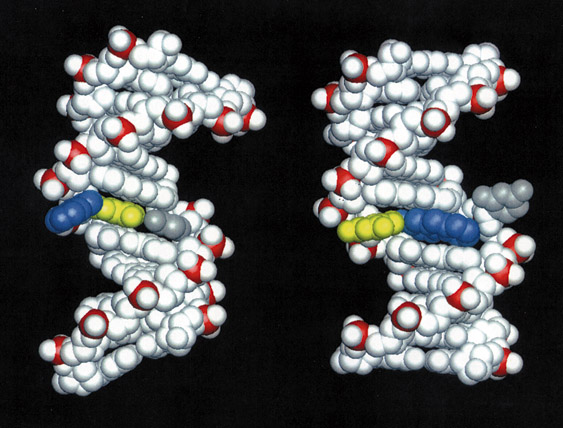
DNA damaged by carcinogenic 2-aminofluorene.
Structures of DNA damaged by the carcinogenic aromatic amine 2-aminofluorene (AF). Left: AF in the B-DNA major groove, the predominant structure at a mutational coldspot. Right: AF inserted into the helix with displacement of the damaged guanine, the predominant structure at a mutational hotspot. Color code: AF: blue; AF-damaged guanine: yellow; cytosine partner to damaged guanine: gray. Molecular Understanding of Mutagenicity Brian E. Hingerty, Oak Ridge National Laboratory Suse Broyde, New York University Dinshaw J. Patel, Memorial Sloan Kettering Cancer Center Research Objectives To elucidate why certain DNA base sequences are mutational hotspots when damaged by carcinogenic environmental chemicals. Computational Approach Molecular mechanics calculations in combination with data from NMR experiments in the form of distances between hydrogens on the carcinogen-damaged DNA molecule are employed to produce molecular views of the damaged DNA that are in agreement with the data. The computations are carried out with the molecular mechanics program DUPLEX on the Cray C90. Accomplishments The aromatic amines are a category of environmental carcinogens present in tobacco smoke, automobile exhaust, dyes and other industrial products, and broiled meats and fish. These substances, when activated biochemically, can bind to DNA and subsequently cause a mutation when the DNA replicates. Such mutations are widely believed to be the initiating event in carcinogenesis by these substances. Often, the target base in the DNA to which the carcinogen binds is guanine (G). Interestingly, it has been found that a carcinogen-bound guanine may be highly mutagenic (a hotspot) or weakly or non-mutagenic, depending on what the neighbor bases are. One example of such a sequence that has been of considerable interest comes from the E. coli bacterium. It is known as the NarI sequence and contains the bases G1-G2-C-G3, where C is the base cytosine. Surprisingly, G3 is a mutational hotspot when bound by certain aromatic amine carcinogens while G1 and G2 are not. The underlying reason for this difference has been a mystery and is of great importance because it is a paradigm for mutational hotspots, such as in the p53 gene, which are found mutated in many human tumors. We have elucidated the structure of a DNA duplex containing the NarI sequence linked at G1, G2, or G3 with a model aromatic amine carcinogen known as 2-aminofluorene (AF), using a combination of high-resolution NMR solution studies and molecular mechanics computations. These studies have revealed a striking difference in structure when the carcinogen damage is at G3, compared to G1 or G2. When the AF is at G1 or G2, it resides preponderantly in the major groove of an unperturbed B-DNA double helix. However, when the AF is at G3, it resides half the time in a position where it is inserted into the helix, causing the damaged guanine to be displaced from its normal helix-inserted position. It is plausible that this structural distortion, if also present during DNA replication in the cell, could be responsible for the failure of the DNA to replicate normally when the hotspot is damaged, leading to the mutatagenic consequence. Significance This work is the first delineation of structural distinctions between mutagenic hotspots and coldspots, revealing how subtle differences in base sequence can produce remarkable differences in structure that can explain the hotspot phenomenon. Publications Mao B., Gu Z., Hingerty B. E., Broyde S. and Patel D. J. N. d. Solution structure of the aminofluorene [AF]-intercalated conformer of the syn [AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry, In Press. Mao B., Gu Z., Hingerty B. E., Broyde S. and Patel D. J. N. d. Solution structure of the aminofluorene [AF]-external conformer of the anti [AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry, In Press. |
|
|
|
DNA, antisense
The DNA sense strand looks like the messenger RNA (mRNA) transcript, and can therefore be used to read the expected codon sequence that will ultimately be used during translation (protein synthesis) to build an amino acid sequence and then a protein. For example, the sequence "ATG" within a DNA sense strand corresponds to an "AUG" codon in the mRNA, which codes for the amino acid methionine. However, the DNA sense strand itself is not used as the template for the mRNA; it is the DNA antisense strand which serves as the source for the protein code, because, with bases complementary to the DNA sense strand, it is used as a template for the mRNA. Since transcription results in an RNA product complementary to the DNA template strand, the mRNA is complementary to the DNA antisense strand.
Hence, a base triplet 3′-TAC-5′ in the DNA antisense strand (complementary to the 5′-ATG-3′ of the DNA sense strand) is used as the template which results in a 5′-AUG-3′ base triplet in the mRNA. The DNA sense strand will have the triplet ATG, which looks similar to the mRNA triplet AUG but will not be used to make methionine because it will not be directly used to make mRNA. The DNA sense strand is called a "sense" strand not because it will be used to make protein (it won't be), but because it has a sequence that corresponds directly to the RNA codon sequence. By this logic, the RNA transcript itself is sometimes described as "sense". (W)
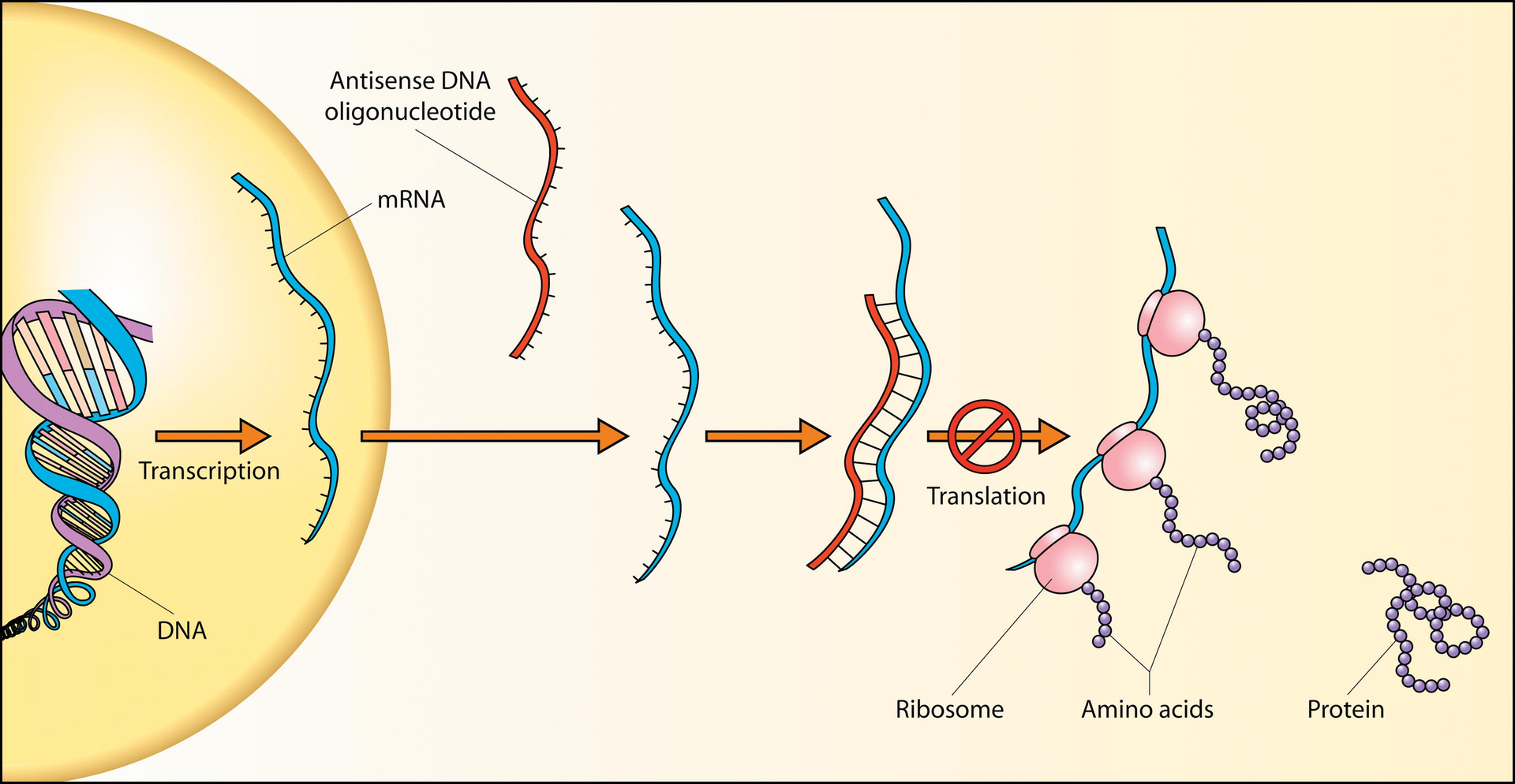
Schematic showing how antisense DNA strands can interfere with protein translation. |
|
|
|
DNA-binding domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. (W)
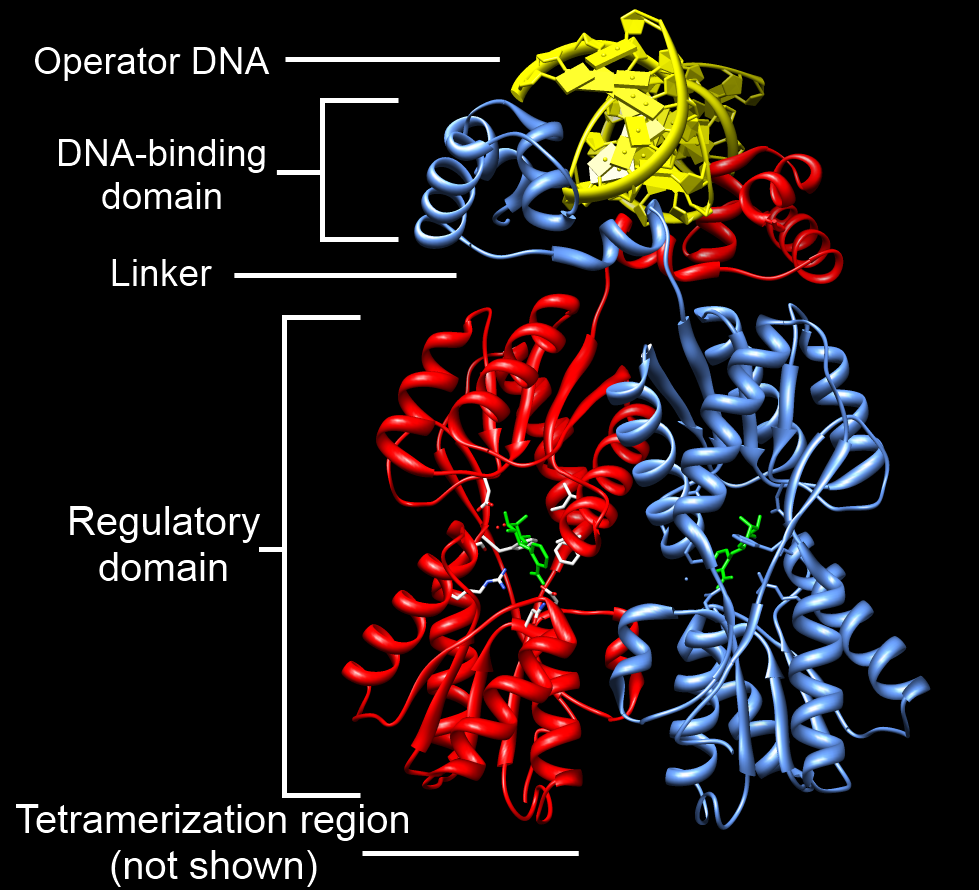
Example of a DNA-binding domain in the context of a protein. The N-terminal DNA-binding domain (labeled) of Lac repressor is regulated by a C-terminal regulatory domain (labeled). The regulatory domain binds an allosteric effector molecule (green). The allosteric response of the protein is communicated from the regulatory domain to the DNA binding domain through the linker region.
Annotated structure of LacI dimer bound to operator DNA and OPNF based on PDB structure 1efa. Created by user SocratesJedi using UCSF Chimera on 2011-10-27. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). |
|
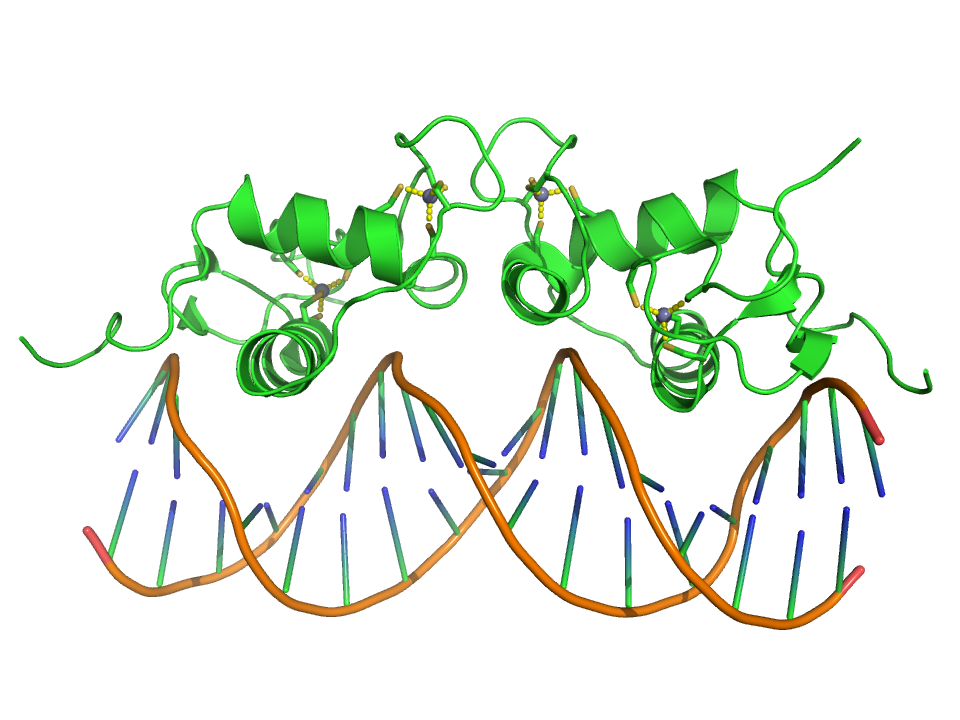
Crystallographic structure (PDB: 1R4O) of a dimer of the zinc finger containing DBD of the glucocorticoid receptor (top) bound to DNA (bottom). Zinc atoms are represented by grey spheres and the coordinating cysteine sidechains are depicted as sticks. |
|
|
|
DNA, chloroplast
Chloroplasts have their own DNA, often abbreviated as cpDNA. It is also known as the plastome when referring to genomes of other plastids. Its existence was first proven in 1962. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, hundreds of chloroplast DNAs from various species have been sequenced, but they are mostly those of land plants and green algae—glaucophytes, red algae, and other algae groups are extremely underrepresented, potentially introducing some bias in views of "typical" chloroplast DNA structure and content. (W)
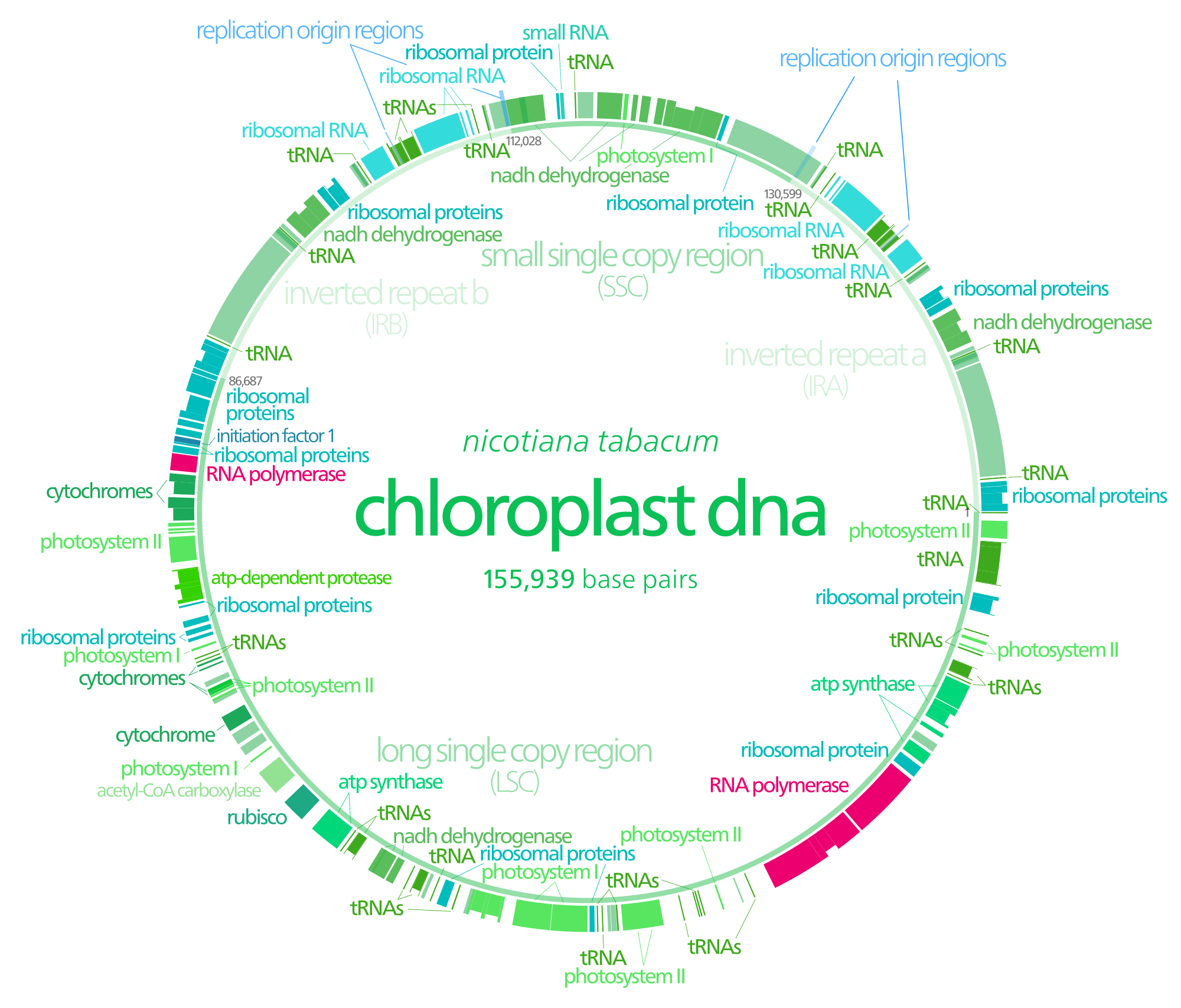
Gene map of tobacco chloroplast DNA. Data was taken from this paper, and input into a Open document spreadsheet to calculate the degree and radian measures, and to generate the SVG data.
Each DNA segment is identified with a path id in the source SVG(albeit not labeled with a visible text object)
Segments with labels on the outside are located on the A strand, segments with labels on the inside are located on the B strand. Segments narrower than the surrounding ones (the notches) indicate introns. Unlabeled faded segments represent open reading frames. (W) |
|
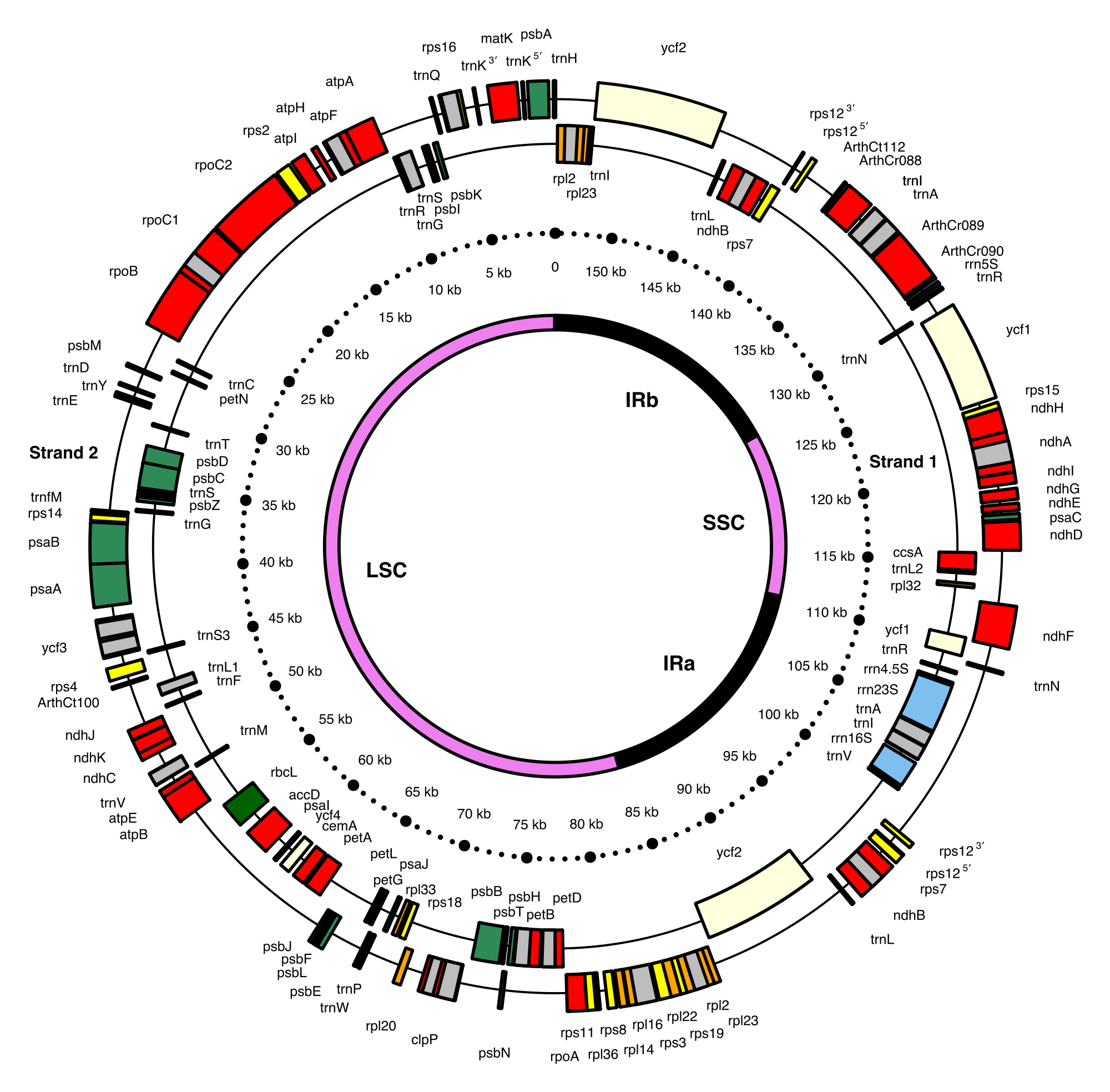
The 154 kb chloroplast DNA map of a model flowering plant (Arabidopsis thaliana: Brassicaceae) showing genes and inverted repeats.
Chloroplast genome map of Arabidopsis thaliana (154,478 bp ; NCBI accession number NC_000932.1 — Sato S., Nakamura Y., Kaneko T., Asamizu E. and Tabata S., 1999. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Research 6 (5), 283-290.). The 136 genes are color coded: small subunit ribosomal proteins (rps, yellow), large subunit ribosomal proteins (rpl, orange), hypothetical chloroplast open reading frames (ycf, lemon), protein-coding genes either involved in photosynthetic reactions (green) or in other functions (red), ribosomal RNAs (rrn, blue), and transfer RNAs (trn, black). Introns are in grey. Some genes consist of 5′ and 3′ portions. Strand 1 and 2 genes are transcribed clockwise and counterclockwise, respectively.
The innermost circle provides the boundaries of the large and small single copy regions (LSC and SSC, violet) separated by a pair of inverted repeats (IRa and IRb, black). (W) |
|
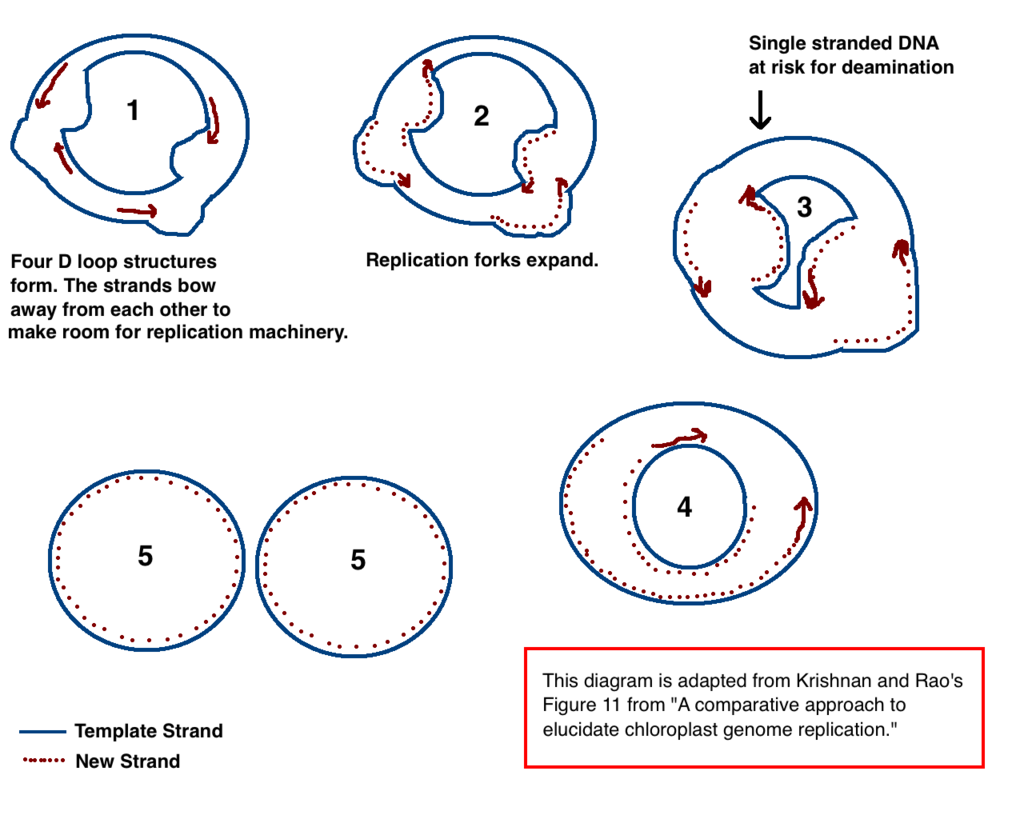
Chloroplast DNA replication via multiple D loop mechanisms. Adapted from Krishnan NM, Rao BJ's paper "A comparative approach to elucidate chloroplast genome replication." |
|
|
|
DNA codon table
List of genetic codes (LINK)
The standard genetic code is traditionally represented as an RNA codon table because, when proteins are made in a cell by ribosomes, it is mRNA that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as "translation table 1." However, with the rise of computational biology and genomics, most genes are now discovered at the DNA level, so a DNA codon table is becoming increasingly useful. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5' → 3' direction. There is the existence of symmetrical and asymmetrical characteristics in genetic codes.
There are 64 different codons in DNA and the below tables; all but three specify an amino acid. These three other codons, deemed stop codons, have specific names: UAG is amber, UGA is opal (sometimes also called umber), and UAA is ochre. Also called "termination" or "nonsense" codons, these sequences signal the release of the nascent polypeptide from the ribosome. Another three codons, which specify an amino acid, are called start codons. The most common start codon is AUG, which is read as methionine. Alternative start codons depending on the organism include "GUG" or "UUG"; these codons normally represent valine and leucine, respectively, but as start codons they are translated as methionine or formylmethionine. These start codons, along with sequences such as an initiation factor, initiate translation.
The first table, the standard table, can be used to translate nucleotide triplets into the corresponding amino acid or the appropriate signal if it is a start or stop codon. The second table, appropriately called the inverse, does the opposite: it can be used to deduce a possible triplet code if the amino acid order is known. (W)
Standard DNA codon table
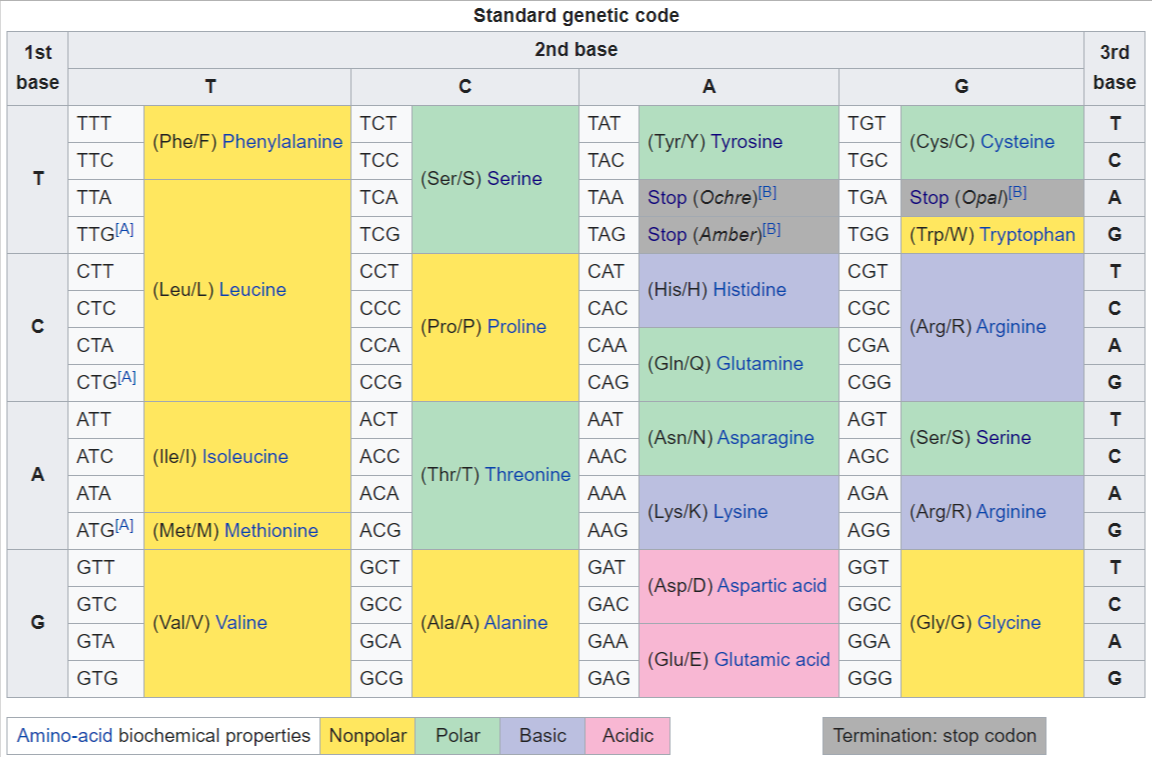
Notes
- A The codon ATG both codes for methionine and serves as an initiation site: the first ATG in an mRNA's coding region is where translation into protein begins. The other start codons listed by GenBank are rare in eukaryotes and generally codes for Met/fMet.
- B ^ ^ ^ The historical basis for designating the stop codons as amber, ochre and opal is described in the autobiography of Sydney Brenner[8] and in a historical article by Bob Edgar.
|
|
|
DNA computing
DNA computing is an emerging branch of computing which uses DNA, biochemistry, and molecular biology hardware, instead of the traditional silicon-based computer technologies. Research and development in this area concerns theory, experiments, and applications of DNA computing. Although the field originally started with the demonstration of a computing application by Len Adleman in 1994, it has now been expanded to several other avenues such as the development of storage technologies, nanoscale imaging modalities, synthetic controllers and reaction networks, etc. (W) |
|
DNA condensation
DNA condensation refers to the process of compacting DNA molecules in vitro or in vivo. Mechanistic details of DNA packing are essential for its functioning in the process of gene regulation in living systems. Condensed DNA often has surprising properties, which one would not predict from classical concepts of dilute solutions. Therefore, DNA condensation in vitro serves as a model system for many processes of physics, biochemistry and biology. In addition, DNA condensation has many potential applications in medicine and biotechnology.
DNA diameter is about 2 nm, while the length of a stretched single molecule may be up to several dozens of centimetres depending on the organism. Many features of the DNA double helix contribute to its large stiffness, including the mechanical properties of the sugar-phosphate backbone, electrostatic repulsion between phosphates (DNA bears on average one elementary negative charge per each 0.17 nm of the double helix), stacking interactions between the bases of each individual strand, and strand-strand interactions. DNA is one of the stiffest natural polymers, yet it is also one of the longest molecules. This means that at large distances DNA can be considered as a flexible rope, and on a short scale as a stiff rod. Like a garden hose, unpacked DNA would randomly occupy a much larger volume than when it is orderly packed. Mathematically, for a non-interacting flexible chain randomly diffusing in 3D, the end-to-end distance would scale as a square root of the polymer length. For real polymers such as DNA, this gives only a very rough estimate; what is important, is that the space available for the DNA in vivo is much smaller than the space that it would occupy in the case of a free diffusion in the solution. To cope with volume constraints, DNA can pack itself in the appropriate solution conditions with the help of ions and other molecules. Usually, DNA condensation is defined as "the collapse of extended DNA chains into compact, orderly particles containing only one or a few molecules".(3) This definition applies to many situations in vitro and is also close to the definition of DNA condensation in bacteria as "adoption of relatively concentrated, compact state occupying a fraction of the volume available".(4) In eukaryotes, the DNA size and the number of other participating players are much larger, and a DNA molecule forms millions of ordered nucleoprotein particles, the nucleosomes, which is just the first of many levels of DNA packing.
- Bloomfield, VA (1997). "DNA condensation by multivalent cations". Biopolymers. 44 (3): 269–82. CiteSeerX 10.1.1.475.3765. doi:10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. PMID 9591479.
- ^ Zimmerman, SB; Murphy, LD (1996). "Macromolecular crowding and the mandatory condensation of DNA in bacteria". FEBS Letters. 390 (3): 245–8. doi:10.1016/0014-5793(96)00725-9. PMID 8706869.
(W)
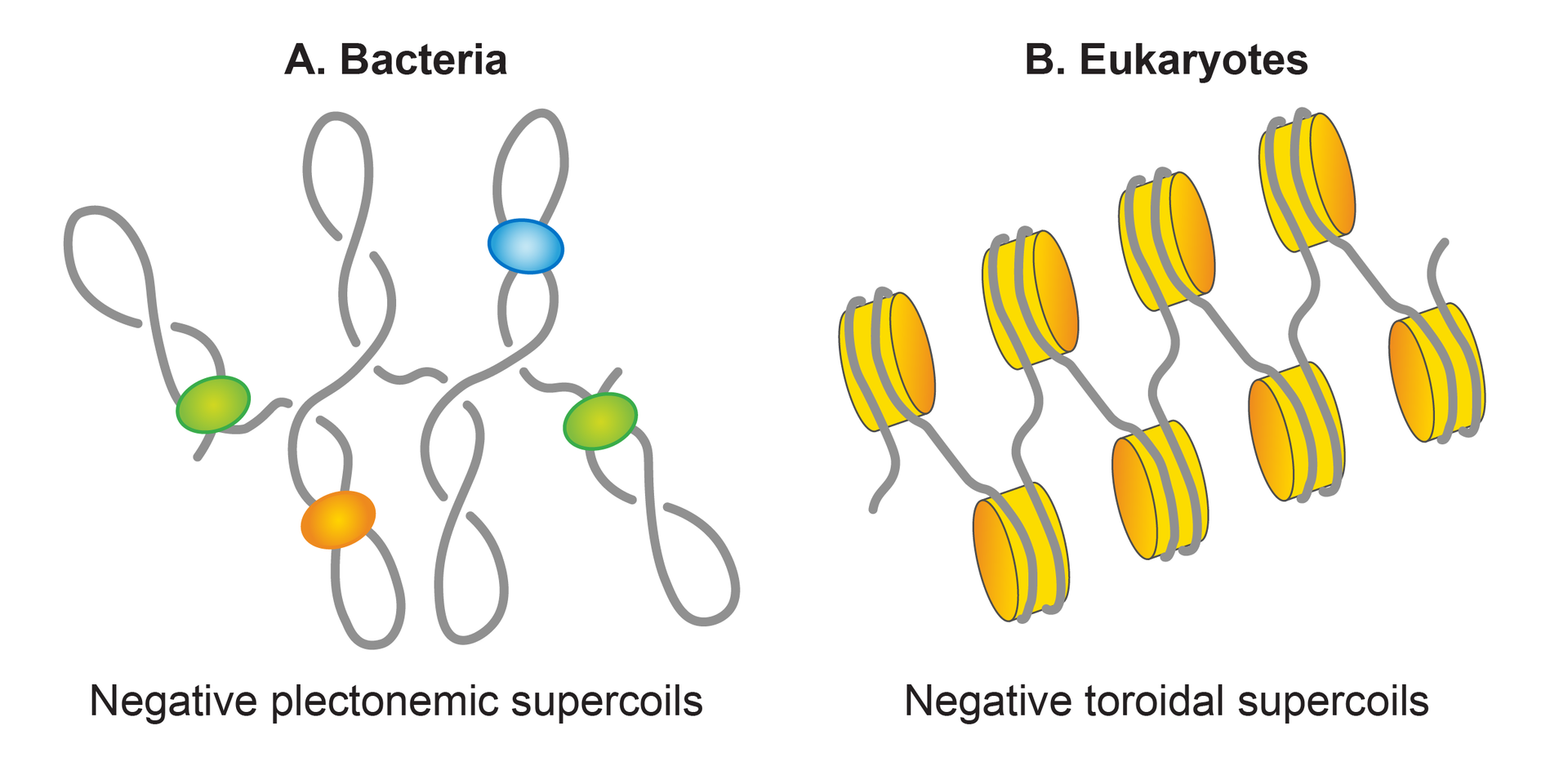
Basic units of genomic organization in bacteria and eukaryotes.
Basic units of genomic organization in bacteria and eukaryotes Genomic DNA, depicted as a grey line, is negatively supercoiled in both bacteria and eukaryotes. However, the negatively supercoiled DNA is organized in the plectonemic form in bacteria, whereas it is organized in the toroidal form in eukaryotes. Nucleoid associated proteins (NAPs), shown as colored spheres, restrain half of the plectonemic supercoils, whereas almost all of the toroidal supercoils are induced as well as restrained by nucleosomes (colored orange), formed by wrapping of DNA around histones. |
|
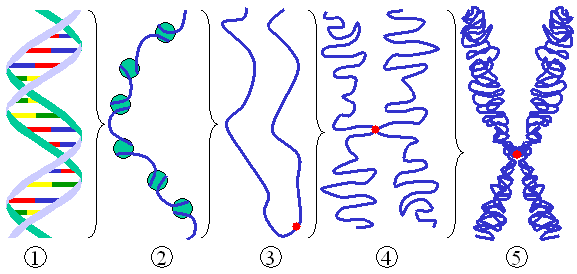
Different levels of DNA condensation. (1) Single DNA strand. (2) Chromatin strand (DNA with histones). (3) Chromatin during interphase with centromere. (4) Condensed chromatin during prophase. (Two copies of the DNA molecule are now present) (5) Chromosome during metaphase. |
|
|
|
DNA damage (naturally occurring)
DNA damage is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of standard base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly.
DNA damage and mutation have different biological consequences. While most DNA damages can undergo DNA repair, such repair is not 100% efficient. Un-repaired DNA damages accumulate in non-replicating cells, such as cells in the brains or muscles of adult mammals, and can cause aging. (Also see DNA damage theory of aging.) In replicating cells, such as cells lining the colon, errors occur upon replication past damages in the template strand of DNA or during repair of DNA damages. These errors can give rise to mutations or epigenetic alterations. Both of these types of alteration can be replicated and passed on to subsequent cell generations. These alterations can change gene function or regulation of gene expression and possibly contribute to progression to cancer.
Throughout the cell cycle there are various checkpoints to ensure the cell is in good condition to progress to mitosis. The three main checkpoints are at G1/s, G2/m, and at the spindle assembly checkpoint regulating progression through anaphase. G1 and G2 checkpoints involve scanning for damaged DNA. During S phase the cell is more vulnerable to DNA damage than any other part of the cell cycle. G2 checkpoint checks for damaged DNA and DNA replication completeness. DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a base missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage. (W)
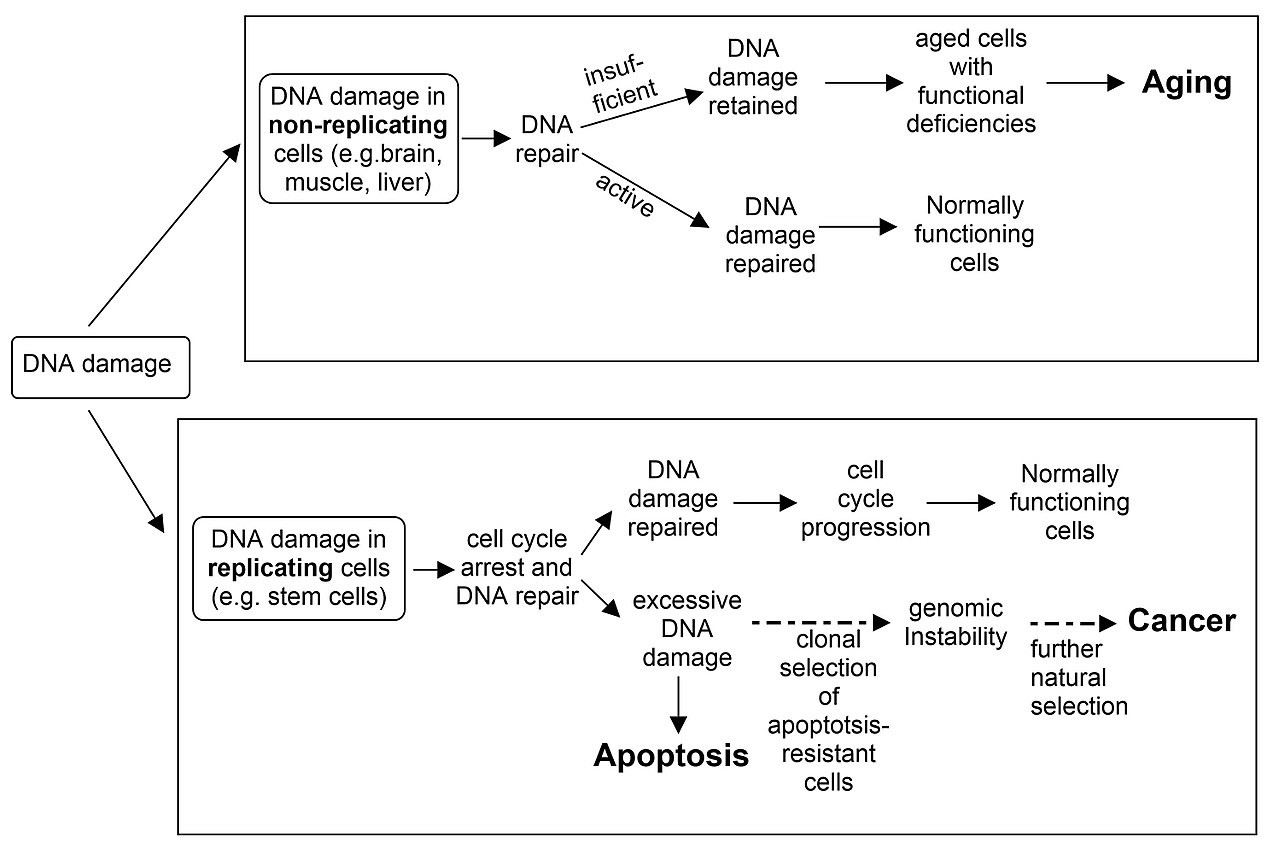
DNA damage in non-replicating cells, if not repaired and accumulated can lead to aging. DNA damage in replicating cells, if not repaired can lead to either apoptosis or to cancer. |
|
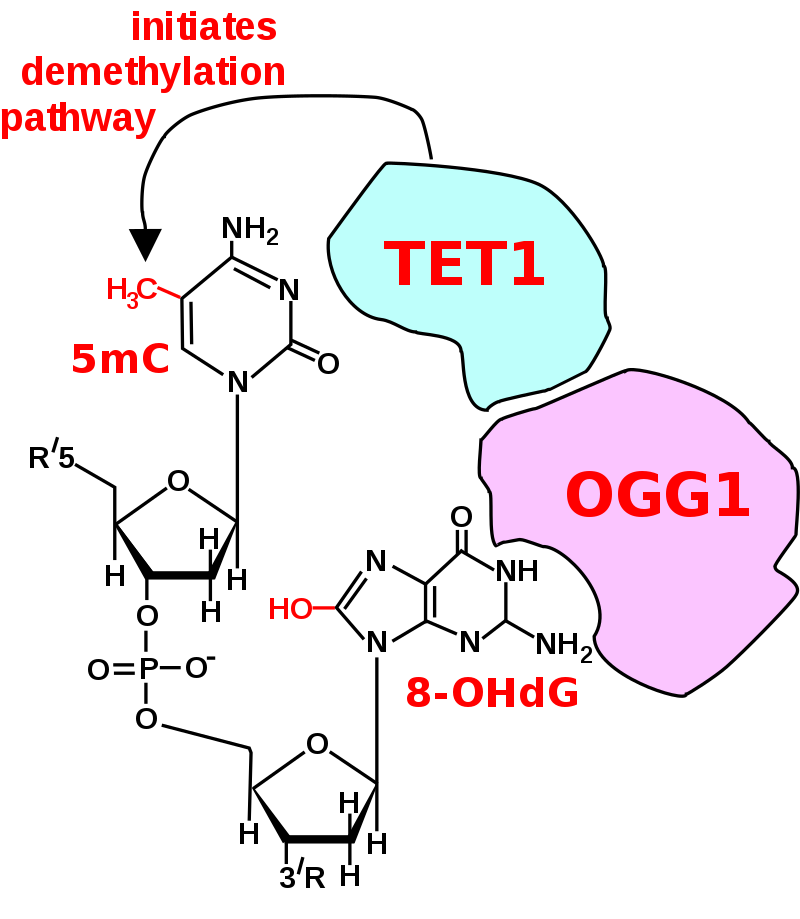
Initiation of DNA demethylation at a CpG site. In adult somatic cells DNA methylation typically occurs in the context of CpG dinucleotides (CpG sites), forming 5-methylcytosine-pG, or 5mCpG. Reactive oxygen species (ROS) may attack guanine at the dinucleotide site, forming 8-hydroxy-2'-deoxyguanosine (8-OHdG), and resulting in a 5mCp-8-OHdG dinucleotide site. The base excision repair enzyme OGG1 targets 8-OHdG and binds to the lesion without immediate excision. OGG1, present at a 5mCp-8-OHdG site recruits TET1 and TET1 oxidizes the 5mC adjacent to the 8-OHdG. This initiates demethylation of 5mC. |
|
|
|
DNA damage theory of aging
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of naturally occurring DNA damages. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrial and nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly (by increasing apoptosis or cellular senescence) or directly (by increasing cell dysfunction). (W) |
|
DNA digital data storage
DNA digital data storage is the process of encoding and decoding binary data to and from synthesized strands of DNA.
While DNA as a storage medium has enormous potential because of its high storage density, its practical use is currently severely limited because of its high cost and very slow read and write times.
In June 2019, scientists reported that all 16 GB of text from Wikipedia's English-language version have been encoded into synthetic DNA. (W)
|
|
DNA-directed RNA interference
DNA-directed RNA interference (ddRNAi) is a gene-silencing technique that utilizes DNA constructs to activate an animal cell's endogenous RNA interference (RNAi) pathways. DNA constructs are designed to express self-complementary double-stranded RNAs, typically short-hairpin RNAs (shRNA), that once processed bring about silencing of a target gene or genes. Any RNA, including endogenous mRNAs or viral RNAs, can be silenced by designing constructs to express double-stranded RNA complementary to the desired mRNA target.
This mechanism has great potential as a novel therapeutic to silence disease-causing genes. Proof-of-concept has been demonstrated across a range of disease models, including viral diseases such as HIV, hepatitis B or hepatitis C, or diseases associated with altered expression of endogenous genes such as drug-resistant lung cancer, neuropathic pain, advanced cancer and retinitis pigmentosa. (W)
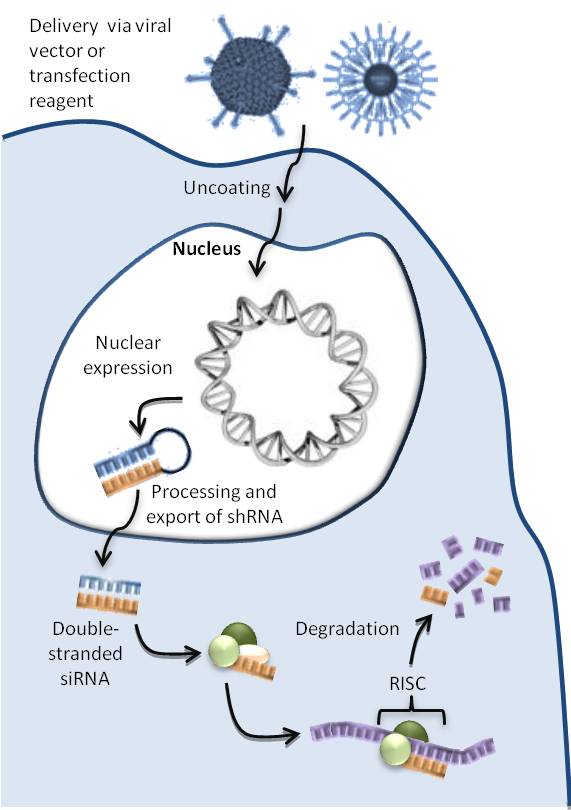
Figure 1: ddRNAi mechanism of action. |
|
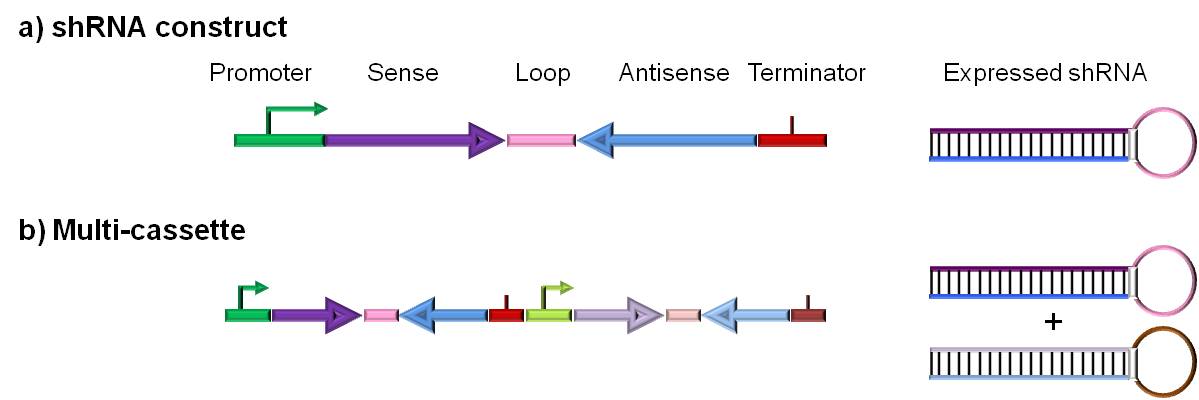
Figure 2: ddRNAi DNA constructs.
Aligned ddRNAi construct designs for the production of therapeutic shRNAs |
|
|
|
DNA ends (Sticky and blunt ends)
DNA ends refer to the properties of the end of DNA molecules, which may be sticky or blunt based on the enzyme which cuts the DNA. The restriction enzyme belong to a larger class of enzymes called exonucleases and endonucleases. Exonucleases remove nucleotide from ends whereas endonuclease cuts at specific position within the DNA.
The concept is used in molecular biology, especially in cloning or when subcloning inserts DNA into vector DNA. Such ends may be generated by restriction enzymes that cut the DNA – a staggered cut generates two sticky ends, while a straight cut generates blunt ends.
Single-stranded DNA molecules
A single-stranded non-circular DNA molecule has two non-identical ends, the 3' end and the 5' end (usually pronounced "three prime end" and "five prime end"). The numbers refer to the numbering of carbon atoms in the deoxyribose, which is a sugar forming an important part of the backbone of the DNA molecule. In the backbone of DNA the 5' carbon of one deoxyribose is linked to the 3' carbon of another by a phosphodiester bond linkage. The 5' carbon of this deoxyribose is again linked to the 3' carbon of the next, and so forth.
Variations in double-stranded molecules
When a molecule of DNA is double stranded, as DNA usually is, the two strands run in opposite directions. Therefore, one end of the molecule will have the 3' end of strand 1 and the 5' end of strand 2, and vice versa in the other end. However, the fact that the molecule is two stranded allows numerous different variations.
Blunt ends
The simplest DNA end of a double stranded molecule is called a blunt end. Blunt end otherwise called as non cohesive restriction enzyme. In a blunt-ended molecule both strands terminate in a base pair. Blunt ends are not always desired in biotechnology since when using a DNA ligase to join two molecules into one, the yield is significantly lower with blunt ends. When performing subcloning, it also has the disadvantage of potentially inserting the insert DNA in the opposite orientation desired. On the other hand, blunt ends are always compatible with each other. Here is an example of a small piece of blunt-ended DNA:
5'-GATCTGACTGATGCGTATGCTAGT-3'
3'-GACTAGACTGACTACGCATACGATCA-5'
Overhangs and sticky ends
Non-blunt ends are created by various overhangs. An overhang is a stretch of unpaired nucleotides in the end of a DNA molecule. These unpaired nucleotides can be in either strand, creating either 3' or 5' overhangs. These overhangs are in most cases palindromic.
The simplest case of an overhang is a single nucleotide. This is most often adenosine and is created as a 3' overhang by some DNA polymerases. Most commonly this is used in cloning PCR products created by such an enzyme. The product is joined with a linear DNA molecule with a 3' thymine overhang. Since adenine and thymine form a base pair, this facilitates the joining of the two molecules by a ligase, yielding a circular molecule. Here is an example of an A-overhang:
5'-ATCTGACTA-3'
3'-TAGACTGA-5'
Longer overhangs are called cohesive ends or sticky ends. They are most often created by restriction endonucleases when they cut DNA. Very often they cut the two DNA strands four base pairs from each other, creating a four-base 5' overhang in one molecule and a complementary 5' overhang in the other. These ends are called cohesive since they are easily joined back together by a ligase.
For example, these two "sticky" ends are compatible:
5'-ATCTGACT + GATGCGTATGCT-3'
3'-TAGACTGACTACG CATACGA-5'
They can form complementary base pairs in the overhang region:
GATGCGTATGCT-3'
5'-ATCTGACT CATACGA-5'
3'-TAGACTGACTACG
Also, since different restriction endonucleases usually create different overhangs, it is possible to create a plasmid by excising a piece of DNA (using a different enzyme for each end) and then joining it to another DNA molecule with ends trimmed by the same enzymes. Since the overhangs have to be complementary in order for the ligase to work, the two molecules can only join in one orientation. This is often highly desirable in molecular biology. (W)
|
|
DNA ligase
DNA ligase is a specific type of enzyme, a ligase, (EC 6.5.1.1) that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.
DNA ligase is used in both DNA repair and DNA replication (see Mammalian ligases). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see Research applications). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA.. (W)
DNA damage, due to environmental factors and normal metabolic processes inside the cell, occurs at a rate of 1,000 to 1,000,000 molecular lesions per cell per day. A special enzyme, DNA ligase (shown here in color), encircles the double helix to repair a broken strand of DNA. DNA ligase is responsible for repairing the millions of DNA breaks generated during the normal course of a cell's life. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. DNA ligases catalyse the crucial step of joining breaks in duplex DNA during DNA repair, replication and recombination, and require either Adenosine triphosphate (ATP) or Nicotinamide adenine dinucleotide (NAD+) as a cofactor. Shown here is DNA ligase I repairing chromosomal damage. The three visable protein structures are: The DNA binding domain (DBD) which is bound to the DNA minor groove both upstream and downstream of the damaged area. The OB-fold domain (OBD) unwinds the DNA slightly over a span of six base pairs and is generally involved in nucleic acid binding. The Adenylation domain (AdD) contains enzymatically active residues that join the broken nucleotides together by catalyzing the formation of a phosphodiester bond between a phosphate and hydroxyl group. It is likely that all mammalian DNA ligases (Ligases I, III, and IV) have a similar ring-shaped architecture and are able to recognize DNA in a similar manner. (See:Nature Article 2004, PDF). |
|
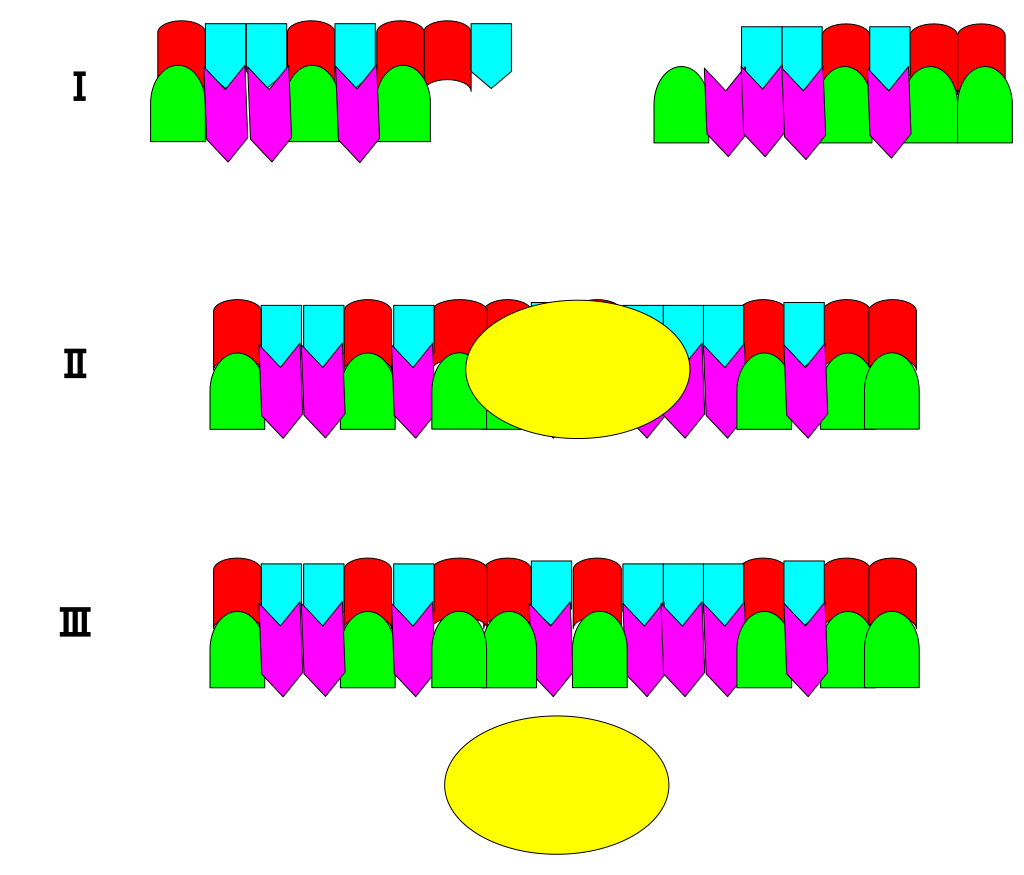
The image demonstrates how ligase (yellow oval) catalyzes two DNA fragment strands. The ligase joins the two fragments of DNA to form a longer strand of DNA by "pasting" them together. |
|
|
|
DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription.In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
Two of DNA's four bases, cytosine and adenine, can be methylated. Cytosine methylation is widespread in both eukaryotes and prokaryotes, even though the rate of cytosine DNA methylation can differ greatly between species: 14% of cytosines are methylated in Arabidopsis thaliana, 4% to 8% in Physarum, 7.6% in Mus musculus, 2.3% in Escherichia coli, 0.03% in Drosophila, 0.006% in Dictyostelium and virtually none (0.0002 to 0.0003%) in Caenorhabditis or fungi such as Saccharomyces cerevisiae and S. pombe (but not N. crassa).:3699 Adenine methylation has been observed in bacterial, plant, and recently in mammalian DNA, but has received considerably less attention. (W)

DNA methylation. |
|
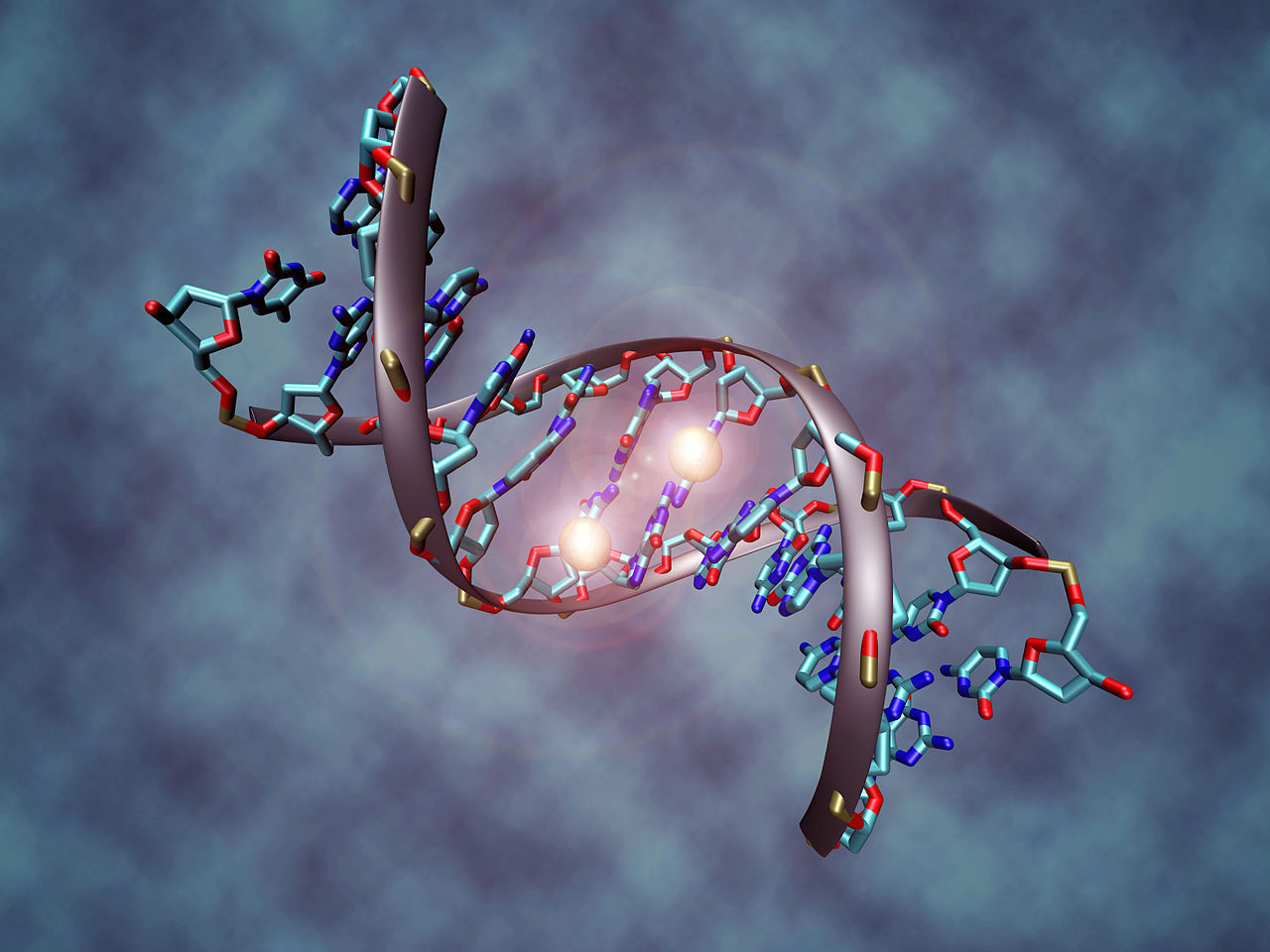
Representation of a DNA molecule that is methylated. The two white spheres represent methyl groups. They are bound to two cytosine nucleotide molecules that make up the DNA sequence.
This image shows a DNA molecule that is methylated on both strands on the center cytosine. DNA methylation plays an important role for epigenetic gene regulation in development and cancer. [Details: The picture shows the crystal structure of a short DNA helix with sequence "accgcCGgcgcc", which is methylated on both strands at the center cytosine. The structure was taken from the Protein Data Bank (accession number 329D), rendering was performed with VMD (Visual Molecular Dynamics rendering program) and post-processing was done in Photoshop. |
|
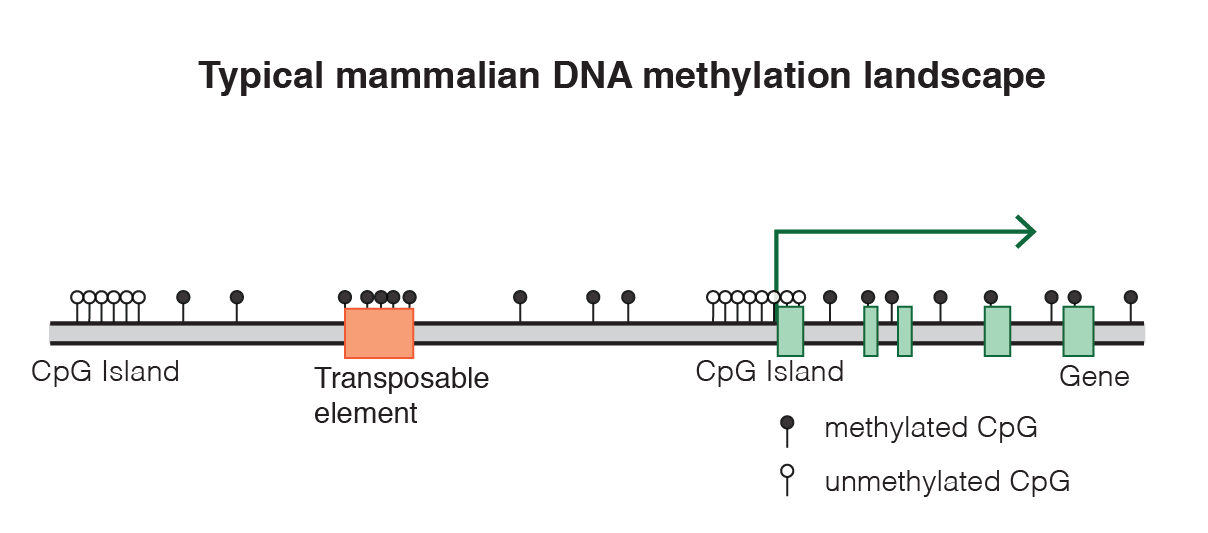
Typical DNA methylation landscape in mammals. |
|
|
|
DNA microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as probes (or reporters or oligos). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called target) under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. (W)
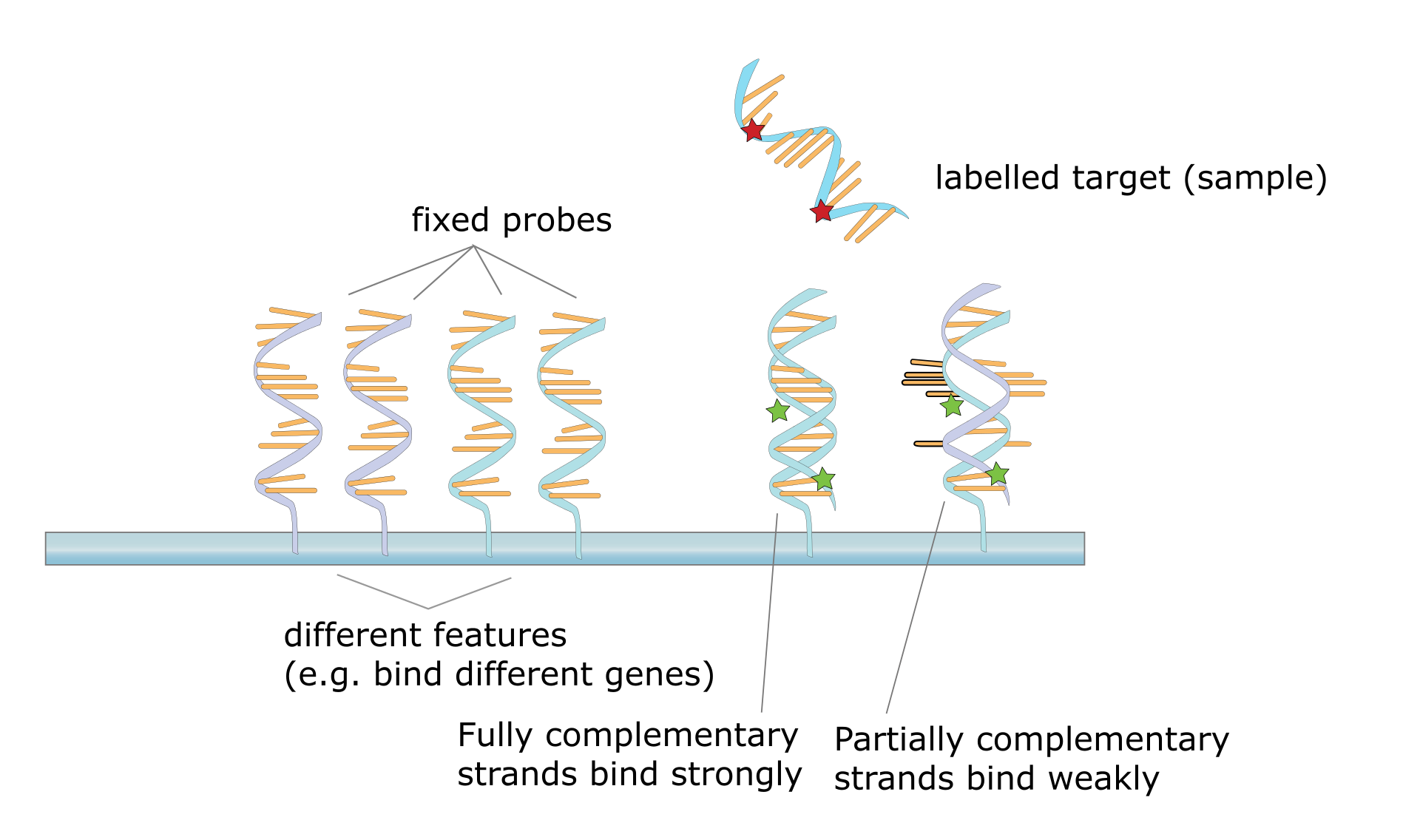
Hybridization of the target to the probe. |
|
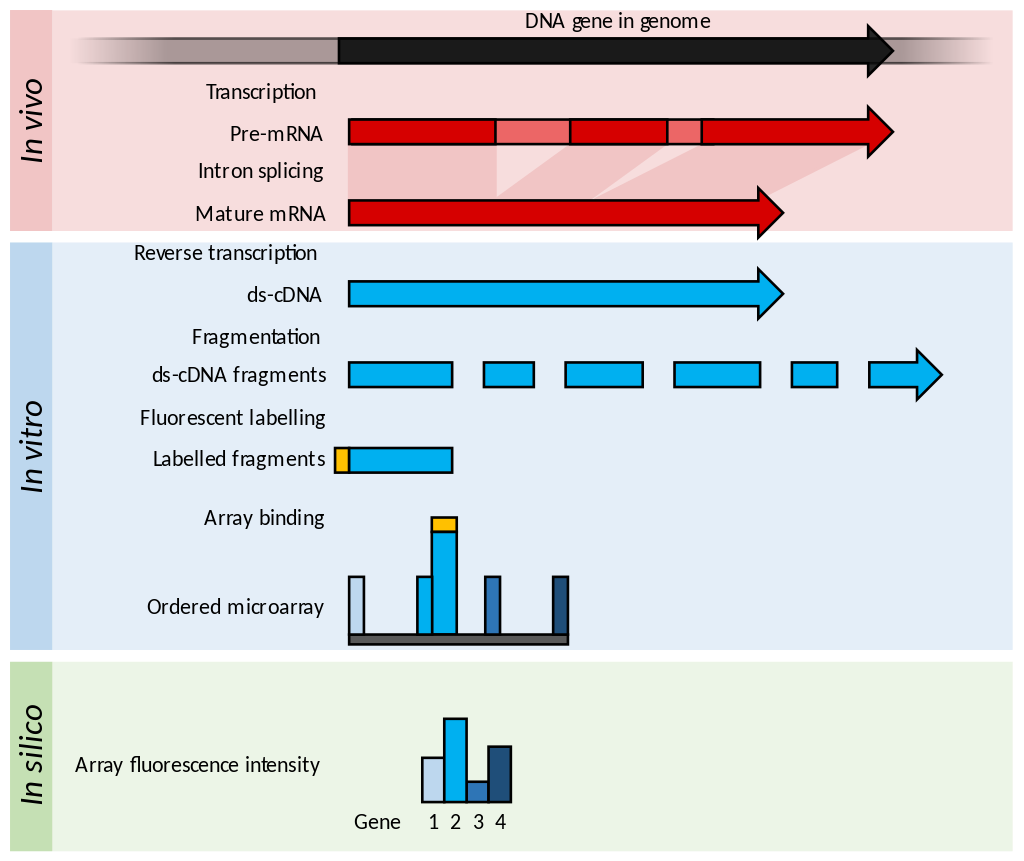
Examples of levels of application of microarrays. Within the organisms, genes are transcribed and spliced to produce mature mRNA transcripts (red). The mRNA is extracted from the organism and reverse transcriptase is used to copy the mRNA into stable ds-cDNA (blue). In microarrays, the ds-cDNA is fragmented and fluorescently labelled (orange). The labelled fragments bind to an ordered array of complementary oligonucleotides, and measurement of fluorescent intensity across the array indicates the abundance of a predetermined set of sequences. These sequences are typically specifically chosen to report on genes of interest within the organism's genome. |
|
|
|
DNA mismatch repair
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage.
Mismatch repair is strand-specific. During DNA synthesis the newly synthesised (daughter) strand will commonly include errors. In order to begin repair, the mismatch repair machinery distinguishes the newly synthesised strand from the template (parental). In gram-negative bacteria, transient hemimethylation distinguishes the strands (the parental is methylated and daughter is not). However, in other prokaryotes and eukaryotes, the exact mechanism is not clear. It is suspected that, in eukaryotes, newly synthesized lagging-strand DNA transiently contains nicks (before being sealed by DNA ligase) and provides a signal that directs mismatch proofreading systems to the appropriate strand. This implies that these nicks must be present in the leading strand, and evidence for this has recently been found. Recent work has shown that nicks are sites for RFC-dependent loading of the replication sliding clamp PCNA, in an orientation-specific manner, such that one face of the donut-shape protein is juxtaposed toward the 3'-OH end at the nick. Loaded PCNA then directs the action of the MutLalpha endonuclease to the daughter strand in the presence of a mismatch and MutSalpha or MutSbeta. (W)
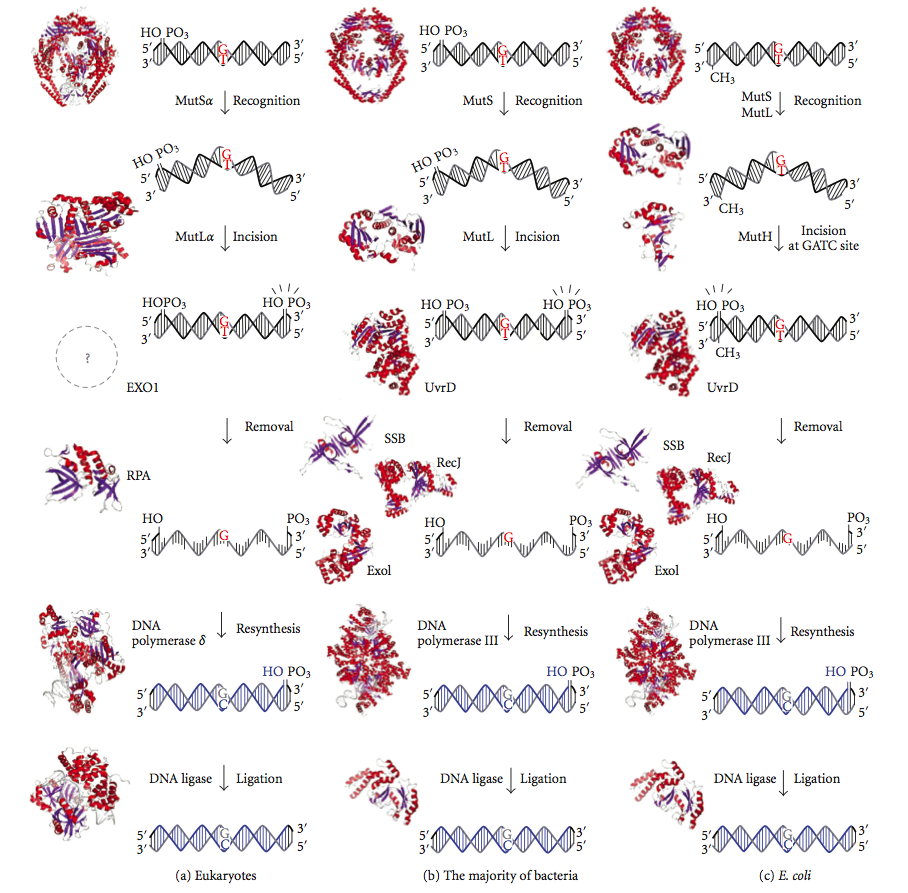
Diagram of DNA mismatch repair pathways. The first column depicts mismatch repair in eukaryotes, while the second depicts repair in most bacteria. The third column shows mismatch repair, to be specific in E. coli. |
|
hpms2-atpgs
Cartoon representation of the molecular structure of protein registered with 1h7u code.
DNA mismatch repair protein, C-terminal domain . |
|
|
|
DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
deoxynucleoside triphosphate + DNAn ⇌ pyrophosphate + DNAn+1.
DNA polymerase adds nucleotides to the three prime (3')-end of a DNA strand, one nucleotide at a time. Every time a cell divides, DNA polymerases are required to duplicate the cell's DNA, so that a copy of the original DNA molecule can be passed to each daughter cell. In this way, genetic information is passed down from generation to generation. (W)
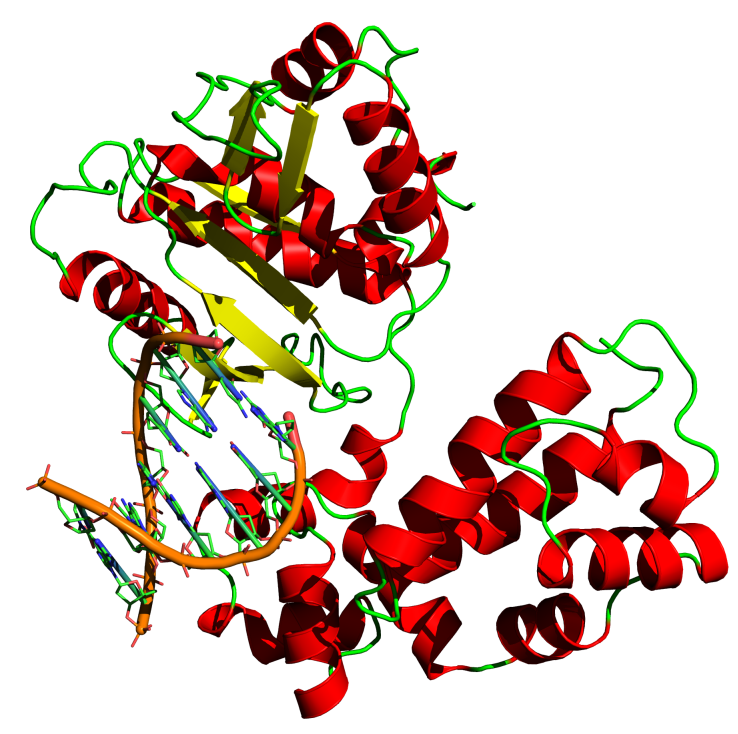
Structure of Homo sapiens DNA polymerase beta, pdb file 7ICG. A bound DNA is also indicated. |
|
DNA polymerase moves along the old strand in the 3'–5' direction, creating a new strand having a 5'–3' direction. |
|
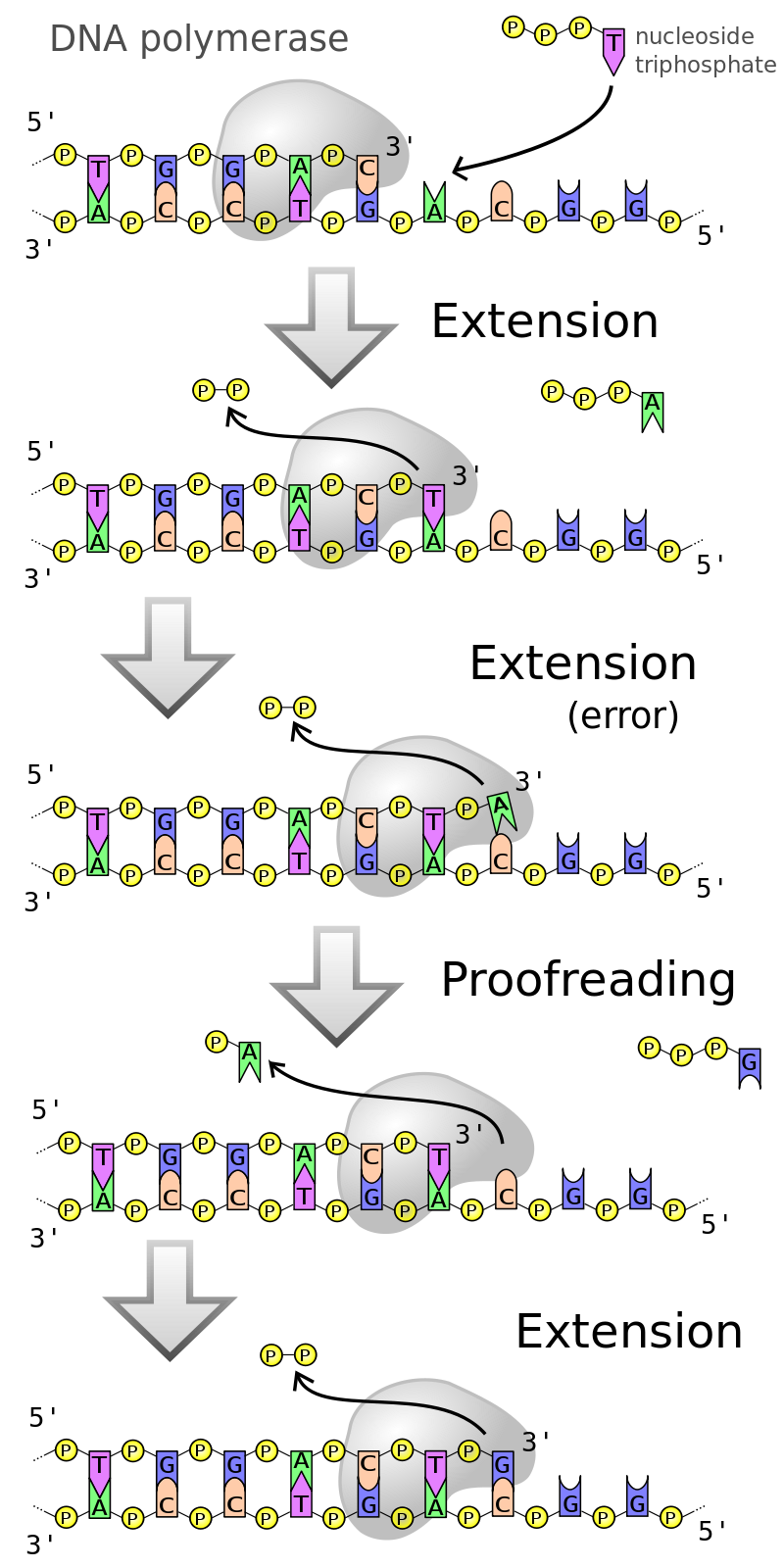
DNA polymerase with proofreading ability. |
|
|
|
DNA primase
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA (or DNA in some organisms) segment called a primer complementary to a ssDNA (single-stranded DNA) template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA. (W)
Asymmetry in the synthesis of leading and lagging strands, with role of DNA primase shown. |
|
Steps in DNA synthesis, with role of DNA primase shown. |
|
|
|
DNA profiling
DNA profiling (also called DNA fingerprinting) is the process of determining an individual's DNA characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding.
DNA profiling is a forensic technique in criminal investigations, comparing criminal suspects' profiles to DNA evidence so as to assess the likelihood of their involvement in the crime. It is also used in parentage testing, to establish immigration eligibility, and in genealogical and medical research. DNA profiling has also been used in the study of animal and plant populations in the fields of zoology, botany, and agriculture. (W)
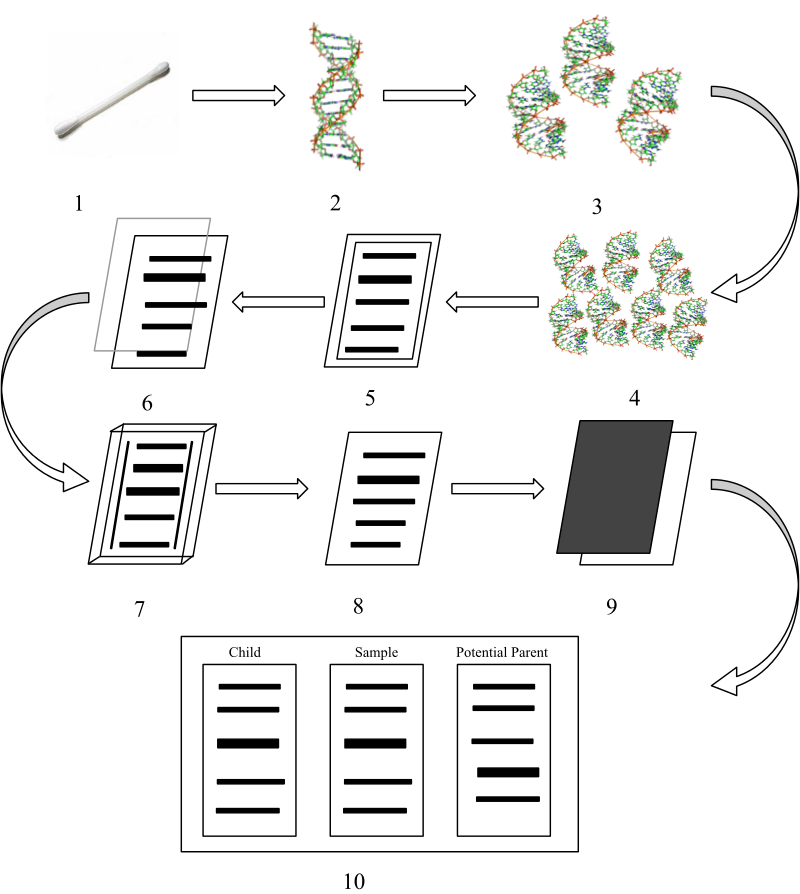
1: A cell sample is taken – usually a cheek swab or blood test 2: DNA is extracted from sample 3: Cleavage of DNA by restriction enzyme – the DNA is broken into small fragments 4: Small fragments are amplified by the polymerase chain reaction – results in many more fragments 5: DNA fragments are separated by electrophoresis 6: The fragments are transferred to an agar plate 7: On the agar plate specific DNA fragments are bound to a radioactive DNA probe 8: The agar plate is washed free of excess probe 9: An x-ray film is used to detect a radioactive pattern 10: The DNA is compared to other DNA samples. |
|
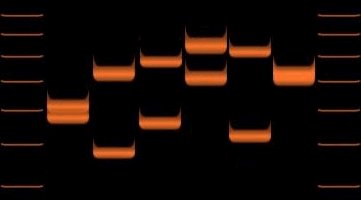
Variations of VNTR allele lengths in 6 individuals. |
|
|
|
DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in as many as 1 million individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.
The rate of DNA repair is dependent on many factors, including the cell type, the age of the cell, and the extracellular environment. A cell that has accumulated a large amount of DNA damage, or one that no longer effectively repairs damage incurred to its DNA, can enter one of three possible states:
- an irreversible state of dormancy, known as senescence
- cell suicide, also known as apoptosis or programmed cell death
- unregulated cell division, which can lead to the formation of a tumor that is cancerous
The DNA repair ability of a cell is vital to the integrity of its genome and thus to the normal functionality of that organism. Many genes that were initially shown to influence life span have turned out to be involved in DNA damage repair and protection.
The 2015 Nobel Prize in Chemistry was awarded to Tomas Lindahl, Paul Modrich, and Aziz Sancar for their work on the molecular mechanisms of DNA repair processes. (W)
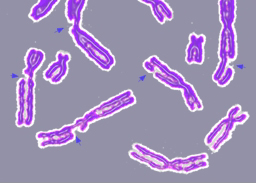
DNA damage resulting in multiple broken chromosomes. |
|
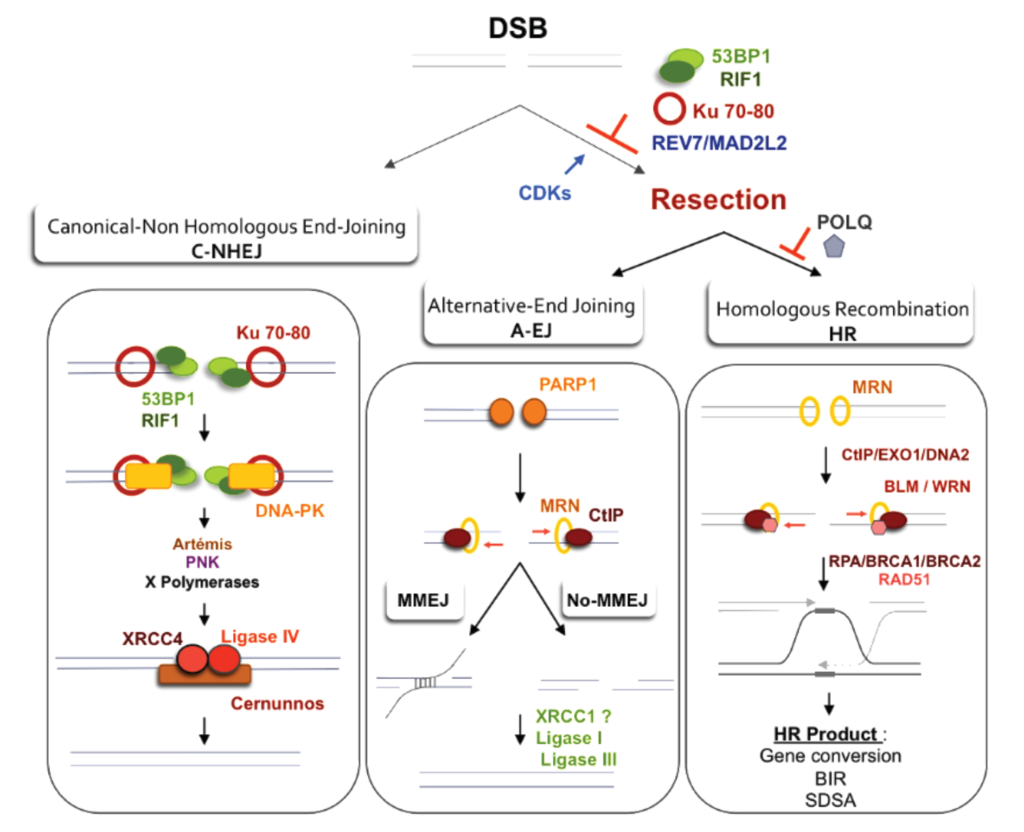
Double-strand break repair pathway models.
(Left panel): Canonical C-NHEJ. The heterodimer Ku80-Ku70 binds to DNA ends, which then recruits DNA-PKcs. In subsequent steps, several proteins including Artemis, polynucleotide kinase (PNK), and members of the polymerase X family process the DNA ends. In the last step, ligase IV associated with its co-factors Xrcc4 and Cernunos/XLF joins the ends. (Right Panel): Resection as a common initiation step for HR and A-EJ at DSB. 53BP1, RIF1 and Ku70-80 heterodimer protect DSB ends from resection and HR and A-EJ actions. The CDK1/2-dependent phosphorylation of CtIP and EXO1 favors the initiation of resection and extension, respectively. Recently, REV7/MAD2L2 was described as an inhibitor of resection and HR, although its role in A-EJ inhibition was not directly studied and remains hypothetical. A short ssDNA resection allows for A-EJ but not homologous recombination, while a long ssDNA resection allows for both A-EJ and HR; however, HR requires the presence of homologous sequences. Recently, POLQ polymerase was shown to inhibit HR and to promote A-EJ at DSBs. A-EJ results in repair that is error-prone and is associated with deletions at the repair junctions with frequent use of microhomologies that are distant from the DSB. Alternative-EJ: Parp1 plays a role in the initiation process, and it has been proposed that a single-strand DNA resection reveals complementary microhomologies (two to four nucleotides or more in length) that can anneal, with gap-filling completing the end-joining. A-EJ is always associated with deletions at the junctions and can involve microhomologies (MMEJ or microhomologies-mediated EJ) that are distant from the DSB. Subsequently, Xrcc1 and ligase III (which can be substituted by ligase I) complete the A-EJ process. Homologous recombination: The first step, which is the initiation of resection, involves the removal of ~50–100 bases of DNA from the 5' end by the MRN complex (Mre11-Rad50-Nbs1) in conjunction with CtIP. The second step, resection extension, is carried out by two alternate pathways involving either the 5' to 3' exonuclease EXO1 or the helicase-topoisomerase complex BLM-TOPIIIα-RMI1-2 in concert with the nuclease CtIP/DNA2. WRN helicase has also been shown to act with CtIP and to stimulate resection in human cells. |
|
DNA ligase, shown above repairing chromosomal damage, is an enzyme that joins broken nucleotides together by catalyzing the formation of an internucleotide ester bond between the phosphate backbone and the deoxyribose nucleotides.
DNA damage, due to environmental factors and normal metabolic processes inside the cell, occurs at a rate of 1,000 to 1,000,000 molecular lesions per cell per day. A special enzyme, DNA ligase (shown here in color), encircles the double helix to repair a broken strand of DNA. DNA ligase is responsible for repairing the millions of DNA breaks generated during the normal course of a cell's life. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. DNA ligases catalyse the crucial step of joining breaks in duplex DNA during DNA repair, replication and recombination, and require either Adenosine triphosphate (ATP) or Nicotinamide adenine dinucleotide (NAD+) as a cofactor. Shown here is DNA ligase I repairing chromosomal damage. The three visable protein structures are: The DNA binding domain (DBD) which is bound to the DNA minor groove both upstream and downstream of the damaged area. The OB-fold domain (OBD) unwinds the DNA slightly over a span of six base pairs and is generally involved in nucleic acid binding. The Adenylation domain (AdD) contains enzymatically active residues that join the broken nucleotides together by catalyzing the formation of a phosphodiester bond between a phosphate and hydroxyl group. It is likely that all mammalian DNA ligases (Ligases I, III, and IV) have a similar ring-shaped architecture and are able to recognize DNA in a similar manner. (See:Nature Article 2004, PDF) . (W) |
|
|
|
Stephen P. Bell (MIT/HHMI) | 1a: Chromosomal DNA Replication | The DNA Replication Fork |
|
|
| |
|
|
|
📹 Stephen P. Bell (MIT/HHMI) | 1a: Chromosomal DNA Replication: The DNA Replication Fork
|
|
Stephen P. Bell (MIT/HHMI) | 1b: Chromosomal DNA Replication | Initiation of DNA Replication |
|
|
| |
|
|
|
📹 Stephen P. Bell (MIT/HHMI) | 1b: Chromosomal DNA Replication | Initiation of DNA Replication
|
|
Stephen P. Bell (MIT/HHMI) | 2: Single-Molecule Studies of Eukaryotic DNA Replication |
|
|
| |
|
|
|
📹 Stephen P. Bell (MIT/HHMI) | 2: Single-Molecule Studies of Eukaryotic DNA Replication
|
|
DNA Replication | MIT 7.01SC Fundamentals of Biology |
|
|
| |
|
|
|
📹 DNA Replication | MIT 7.01SC Fundamentals of Biology
|
|
|
|
DNA replication — Cell cycle regulation
DNA replication is a tightly orchestrated process that is controlled within the context of the cell cycle. Progress through the cell cycle and in turn DNA replication is tightly regulated by the formation and activation of pre-replicative complexes (pre-RCs) which is achieved through the activation and inactivation of cyclin-dependent kinases (Cdks, CDKs). Specifically it is the interactions of cyclins and cyclin dependent kinases that are responsible for the transition from G1 into S-phase.
During the G1 phase of the cell cycle there are low levels of CDK activity. This low level of CDK activity allows for the formation of new pre-RC complexes but is not sufficient for DNA replication to be initiated by the newly formed pre-RCs. During the remaining phases of the cell cycle there are elevated levels of CDK activity. This high level of CDK activity is responsible for initiating DNA replication as well as inhibiting new pre-RC complex formation. Once DNA replication has been initiated the pre-RC complex is broken down. Due to the fact that CDK levels remain high during the S phase, G2, and M phases of the cell cycle no new pre-RC complexes can be formed. This all helps to ensure that no initiation can occur until the cell division is complete.
In addition to cyclin dependent kinases a new round of replication is thought to be prevented through the downregulation of Cdt1. This is achieved via degradation of Cdt1 as well as through the inhibitory actions of a protein known as geminin. Geminin binds tightly to Cdt1 and is thought to be the major inhibitor of re-replication. Geminin first appears in S-phase and is degraded at the metaphase-anaphase transition, possibly through ubiquination by anaphase promoting complex (APC).
Various cell cycle checkpoints are present throughout the course of the cell cycle that determine whether a cell will progress through division entirely. Importantly in replication the G1, or restriction, checkpoint makes the determination of whether or not initiation of replication will begin or whether the cell will be placed in a resting stage known as G0. Cells in the G0 stage of the cell cycle are prevented from initiating a round of replication because the minichromosome maintenance proteins are not expressed. Transition into the S-phase indicates replication has begun. (W)
|
|
DNA replication — Elongation
The formation of the pre-replicative complex (pre-RC) marks the potential sites for the initiation of DNA replication. Consistent with the minichromosome maintenance complex encircling double stranded DNA, formation of the pre-RC does not lead to the immediate unwinding of origin DNA or the recruitment of DNA polymerases. Instead, the pre-RC that is formed during the G1 of the cell cycle is only activated to unwind the DNA and initiate replication after the cells pass from the G1 to the S phase of the cell cycle.
Once the initiation complex is formed and the cells pass into the S phase, the complex then becomes a replisome. The eukaryotic replisome complex is responsible for coordinating DNA replication. Replication on the leading and lagging strands is performed by DNA polymerase ε and DNA polymerase δ. Many replisome factors including Claspin, And1, replication factor C clamp loader and the fork protection complex are responsible for regulating polymerase functions and coordinating DNA synthesis with the unwinding of the template strand by Cdc45-Mcm-GINS complex. As the DNA is unwound the twist number decreases. To compensate for this the writhe number increases, introducing positive supercoils in the DNA. These supercoils would cause DNA replication to halt if they were not removed. Topoisomerases are responsible for removing these supercoils ahead of the replication fork.
The replisome is responsible for copying the entire genomic DNA in each proliferative cell. The base pairing and chain formation reactions, which form the daughter helix, are catalyzed by DNA polymerases. These enzymes move along single-stranded DNA and allow for the extension of the nascent DNA strand by "reading" the template strand and allowing for incorporation of the proper purine nucleobases, adenine and guanine, and pyrimidine nucleobases, thymine and cytosine. Activated free deoxyribonucleotides exist in the cell as deoxyribonucleotide triphosphates (dNTPs). These free nucleotides are added to an exposed 3'-hydroxyl group on the last incorporated nucleotide. In this reaction, a pyrophosphate is released from the free dNTP, generating energy for the polymerization reaction and exposing the 5' monophosphate, which is then covalently bonded to the 3' oxygen. Additionally, incorrectly inserted nucleotides can be removed and replaced by the correct nucleotides in an energetically favorable reaction. This property is vital to proper proofreading and repair of errors that occur during DNA replication. (W)
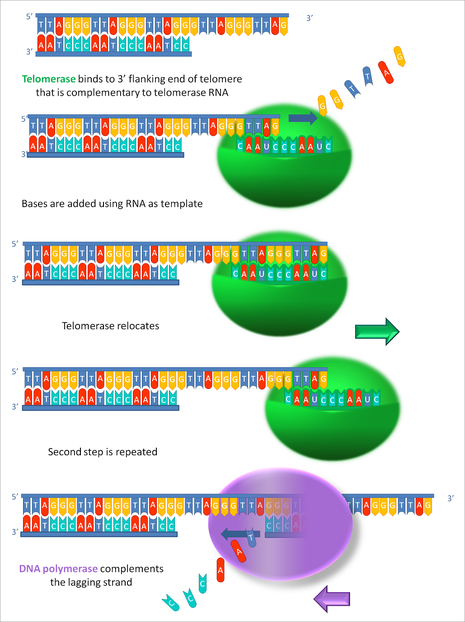
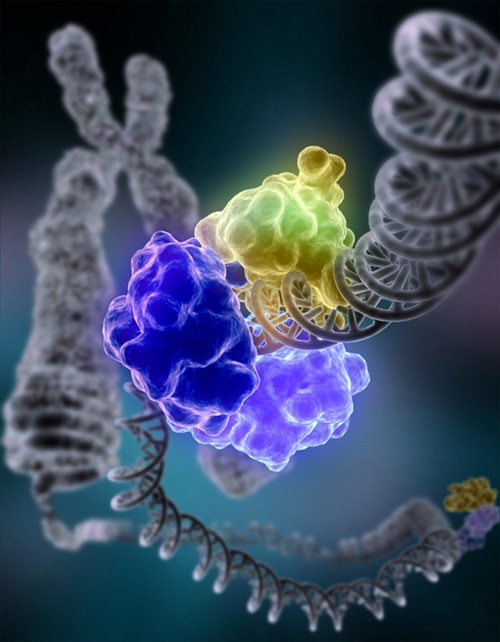
A depiction of telomerase progressively elongating telomeric DNA. |
|
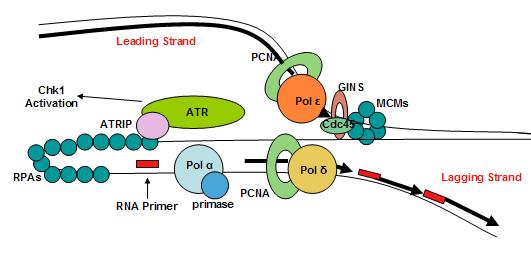
Eukaryotic replisome complex and associated proteins. A loop occurs in the lagging strand. |
|
|
|
DNA replication, eukaryotic
Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.
DNA replication is the action of DNA polymerases synthesizing a DNA strand complementary to the original template strand. To synthesize DNA, the double-stranded DNA is unwound by DNA helicases ahead of polymerases, forming a replication fork containing two single-stranded templates. Replication processes permit the copying of a single DNA double helix into two DNA helices, which are divided into the daughter cells at mitosis. The major enzymatic functions carried out at the replication fork are well conserved from prokaryotes to eukaryotes, but the replication machinery in eukaryotic DNA replication is a much larger complex, coordinating many proteins at the site of replication, forming the replisome.
The replisome is responsible for copying the entirety of genomic DNA in each proliferative cell. This process allows for the high-fidelity passage of hereditary/genetic information from parental cell to daughter cell and is thus essential to all organisms. Much of the cell cycle is built around ensuring that DNA replication occurs without errors.
In G1 phase of the cell cycle, many of the DNA replication regulatory processes are initiated. In eukaryotes, the vast majority of DNA synthesis occurs during S phase of the cell cycle, and the entire genome must be unwound and duplicated to form two daughter copies. During G2, any damaged DNA or replication errors are corrected. Finally, one copy of the genomes is segregated to each daughter cell at mitosis or M phase. These daughter copies each contain one strand from the parental duplex DNA and one nascent antiparallel strand.
This mechanism is conserved from prokaryotes to eukaryotes and is known as semiconservative DNA replication. The process of semiconservative replication for the site of DNA replication is a fork-like DNA structure, the replication fork, where the DNA helix is open, or unwound, exposing unpaired DNA nucleotides for recognition and base pairing for the incorporation of free nucleotides into double-stranded DNA. (W)
|
|
DNA Replication fork — Leading strand, lagging strand
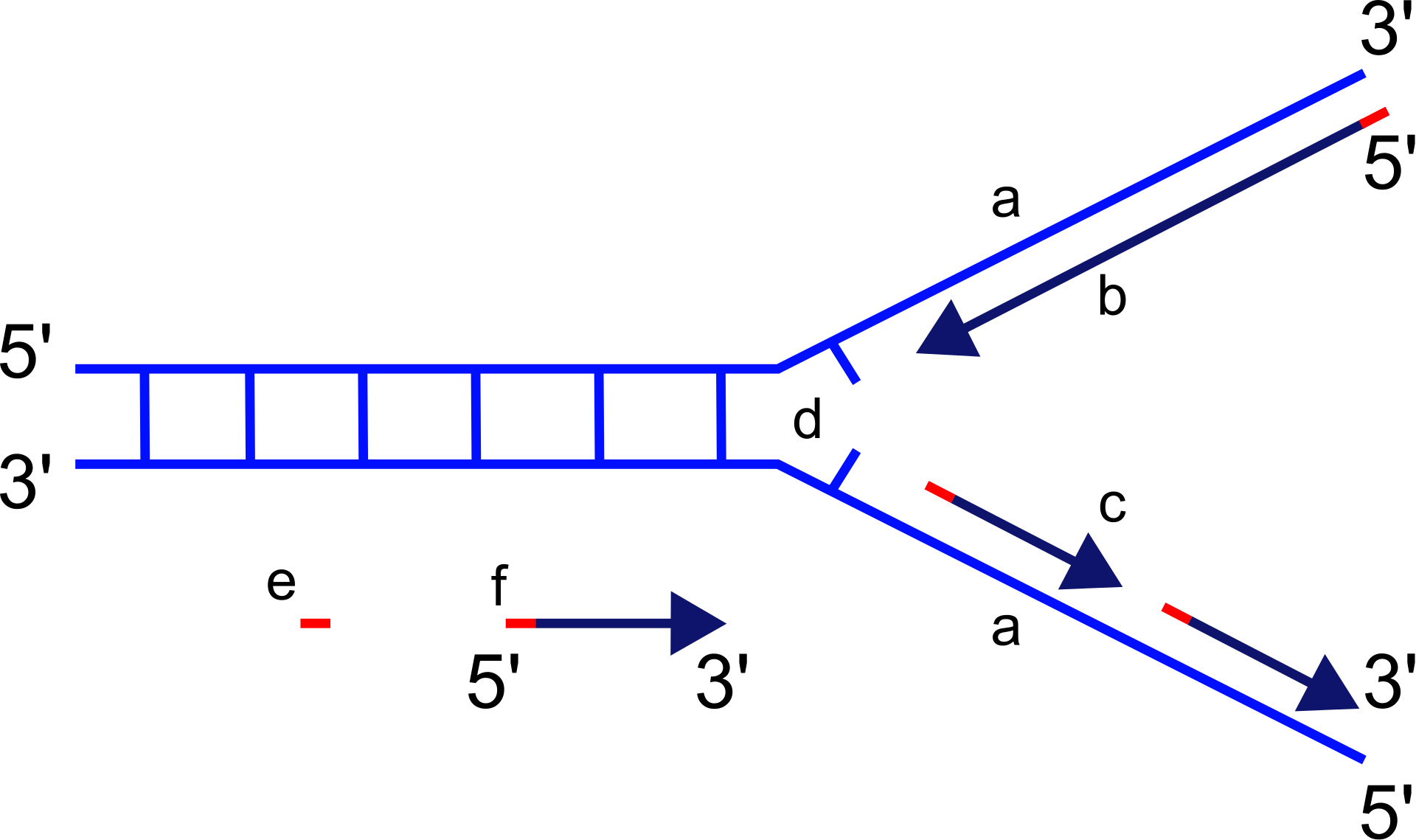
The replication fork is a structure that forms within the long helical DNA during DNA replication. It is created by helicases, which break the hydrogen bonds holding the two DNA strands together in the helix. The resulting structure has two branching "prongs", each one made up of a single strand of DNA. These two strands serve as the template for the leading and lagging strands, which will be created as DNA polymerase matches complementary nucleotides to the templates; the templates may be properly referred to as the leading strand template and the lagging strand template.

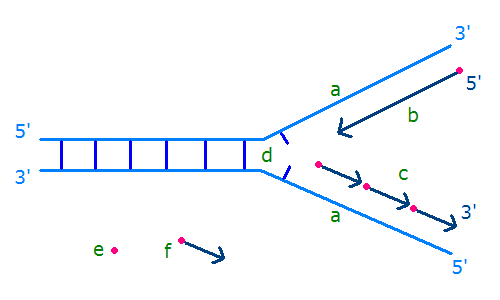
Scheme of the replication fork. a: template, b: leading strand, c: lagging strand, d: replication fork, e: primer, f: Okazaki fragments.
Description: Depiction of DNA replication with replication fork, strands and okazaki-fragments. a: template strands, b: leading strand, c: lagging strand, d: replication fork, e: RNA primer, f: Okazaki fragment. |
|
DNA is read by DNA polymerase in the 3′ to 5′ direction, meaning the nascent strand is synthesized in the 5' to 3' direction. Since the leading and lagging strand templates are oriented in opposite directions at the replication fork, a major issue is how to achieve synthesis of nascent (new) lagging strand DNA, whose direction of synthesis is opposite to the direction of the growing replication fork.
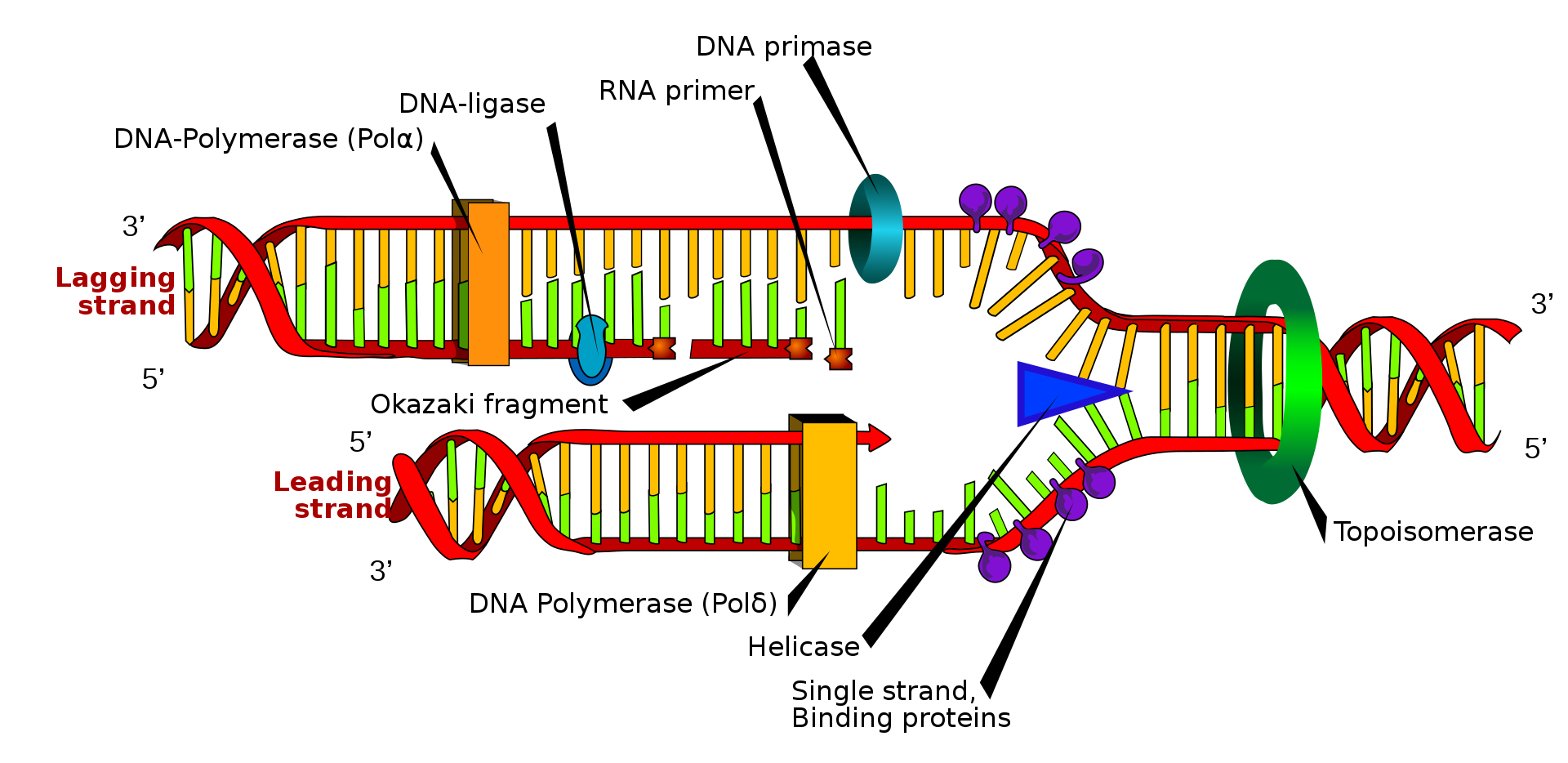
Many enzymes are involved in the DNA replication fork.
DNA replication or DNA synthesis is the process of copying a double-stranded DNA molecule. This process {like many other biological processes} is paramount to all life as we know it. |
|
Leading strand
The leading strand is the strand of nascent DNA which is synthesized in the same direction as the growing replication fork. This sort of DNA replication is continuous.
Lagging strand
The lagging strand is the strand of nascent DNA whose direction of synthesis is opposite to the direction of the growing replication fork. Because of its orientation, replication of the lagging strand is more complicated as compared to that of the leading strand. As a consequence, the DNA polymerase on this strand is seen to "lag behind" the other strand.
The lagging strand is synthesized in short, separated segments. On the lagging strand template, a primase "reads" the template DNA and initiates synthesis of a short complementary RNA primer. A DNA polymerase extends the primed segments, forming Okazaki fragments. The RNA primers are then removed and replaced with DNA, and the fragments of DNA are joined together by DNA ligase. (W)
|
|
DNA replication — Initiation
For a cell to divide, it must first replicate its DNA. DNA replication is an all-or-none process; once replication begins, it proceeds to completion. Once replication is complete, it does not occur again in the same cell cycle. This is made possible by the division of initiation of the pre-replication complex. (W)
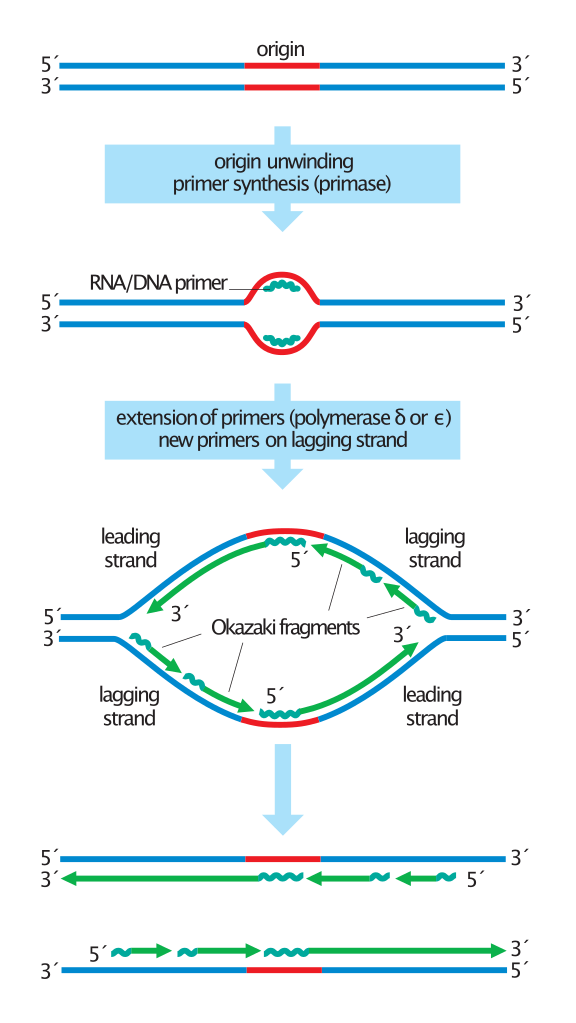
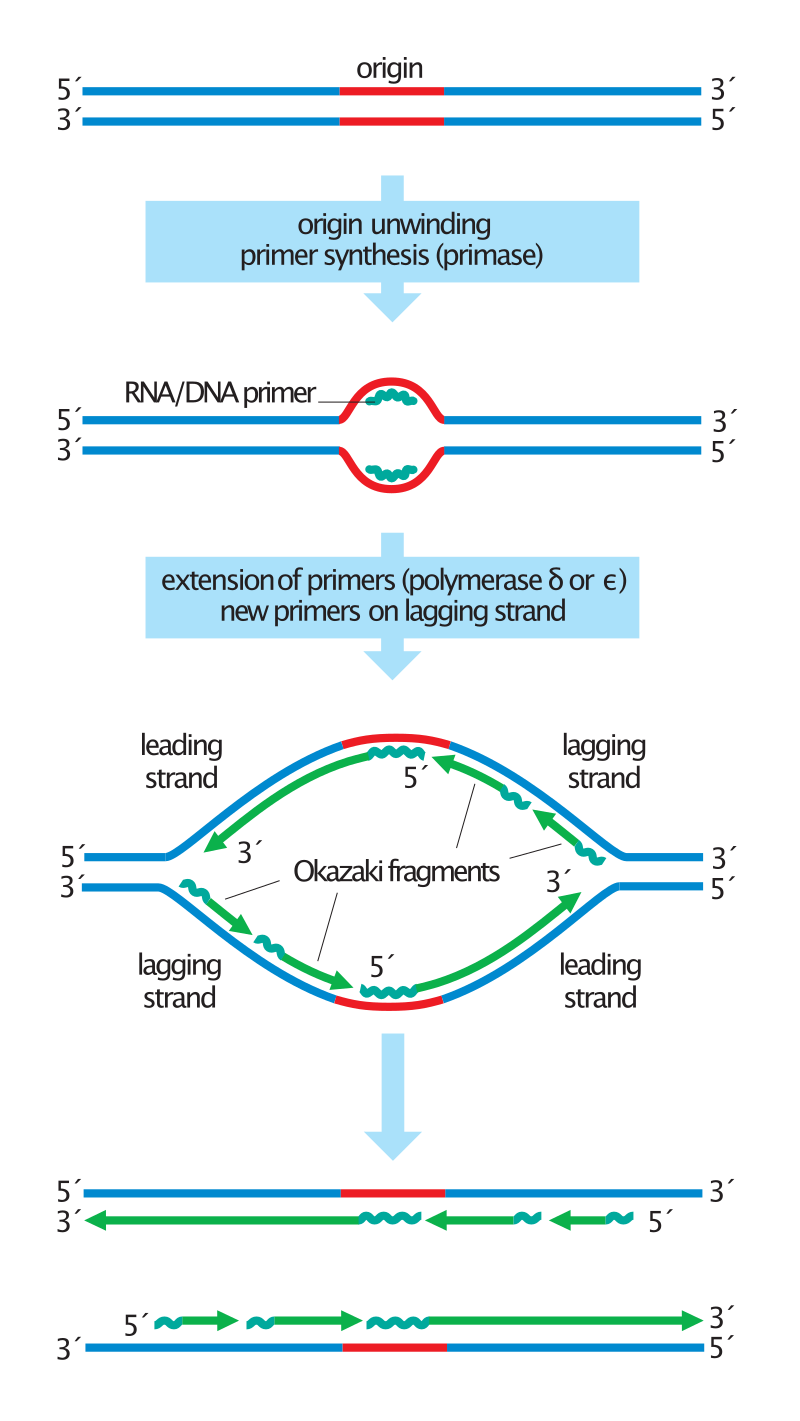
Overview of the steps in DNA replication.
Duplication of the DNA begins with origin unwinding, followed by the synthesis of RNA primers (jagged lines) on both DNA strands. DNA polymerase delta or epsilon extends these primers by adding new DNA (green lines) only in a 5' to 3' direction. On the leading strands, this results in the continuous synthesis of long DNA molecules. Lagging strands, in contrast, are synthesized discontinuously: primers are placed on the template every ~200 nucleotides and extended to form short Okazaki fragments. For simplicity, this diagram does not show the replacement of primers with DNA or the synthesis of telomeres at the chromosome ends. |
|
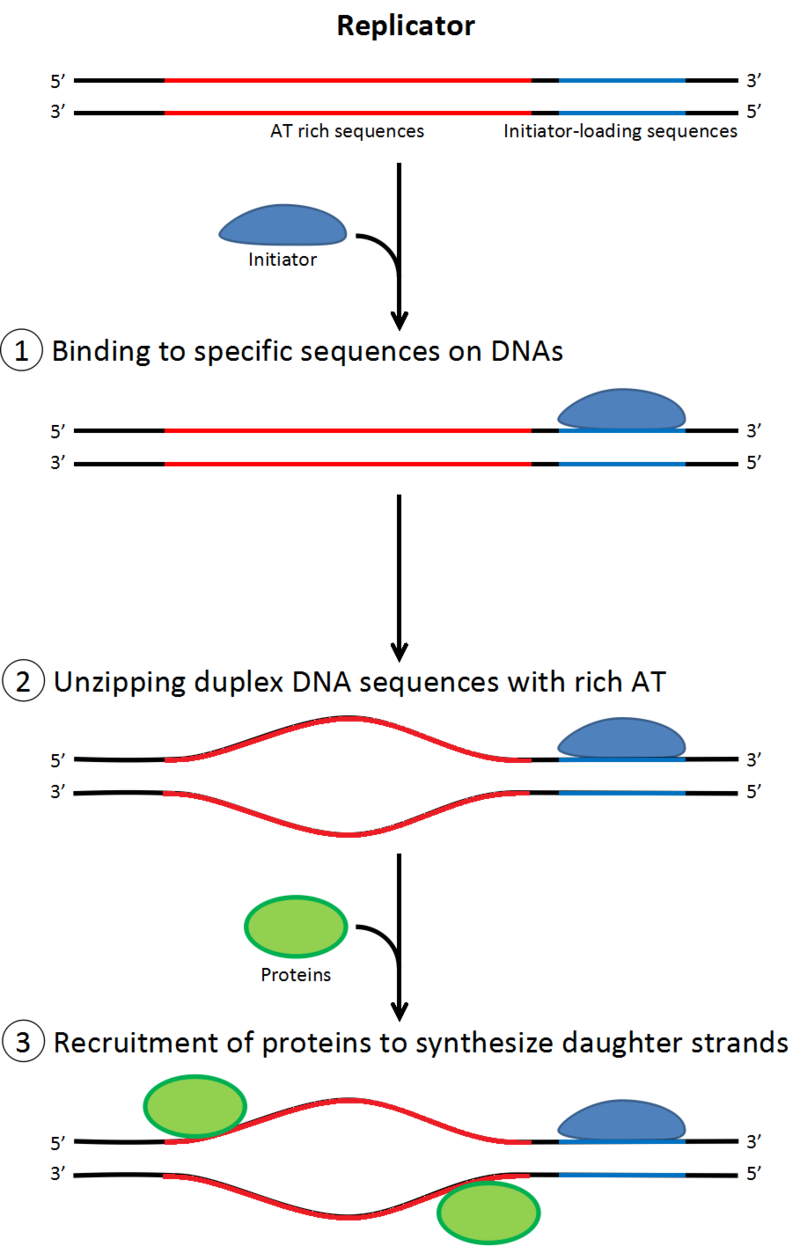
Role of initiators for initiation of DNA replication.
There are AT-rich and initiator-loading sequences on replicators. Initiators bind the initiator-loading sequences and at once the AT-rich sequences are rewound. Initiators recruit other proteins involved in DNA replication. |
|
|
|
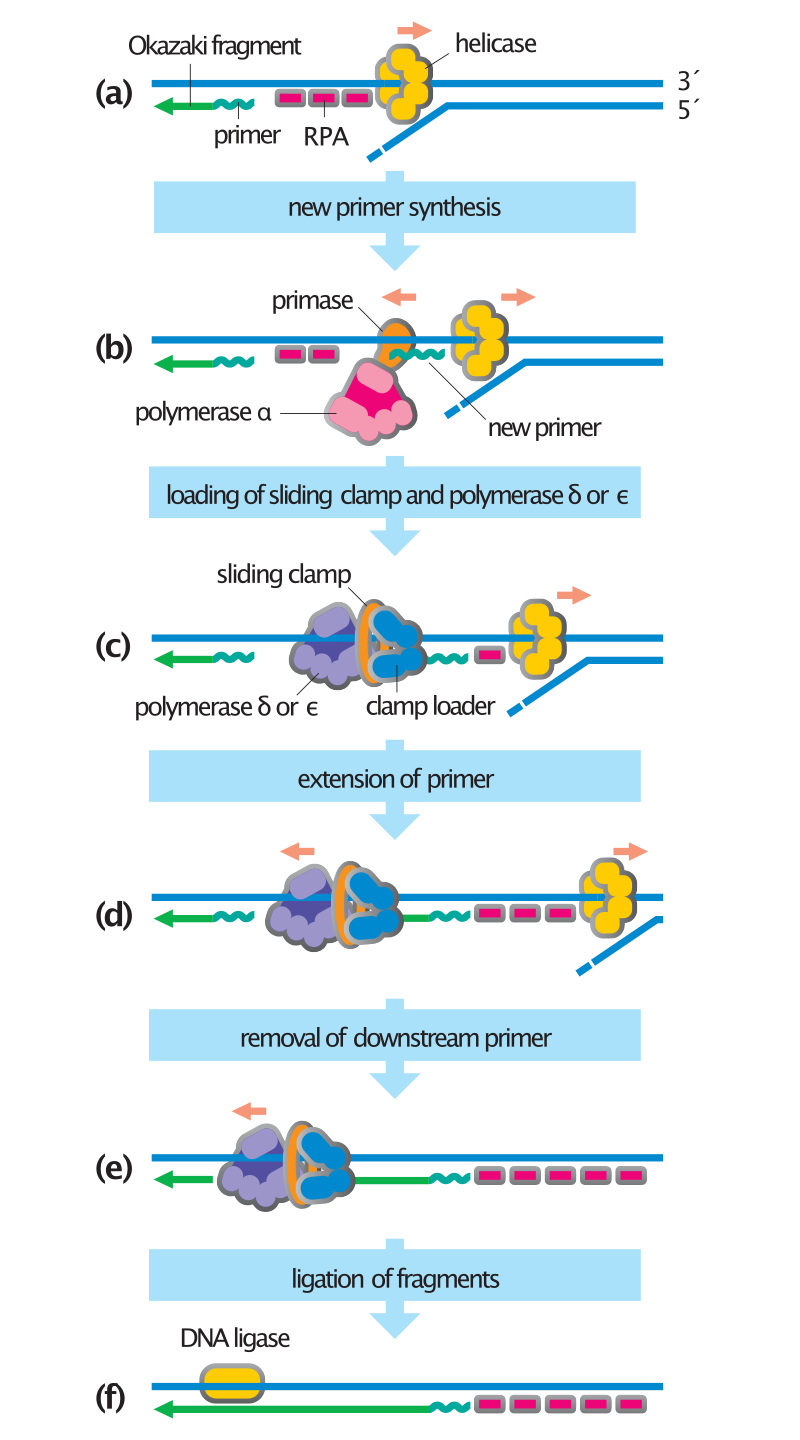
DNA replication — Okazaki fragments
Okazaki fragments are short sequences of DNA nucleotides (approximately 150 to 200 base pairs long in eukaryotes) which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.
During DNA replication, the double helix is unwound and the complementary strands are separated by the enzyme DNA helicase, creating what is known as the DNA replication fork. Following this fork, DNA primase and then DNA polymerase begin to act in order to create a new complementary strand. Because these enzymes can only work in the 5’ to 3’ direction, the two unwound template strands are replicated in different ways. One strand, the leading strand, undergoes a continuous replication process since its template strand has 3’ to 5’ directionality, allowing the polymerase assembling the leading strand to follow the replication fork without interruption. The lagging strand, however, cannot be created in a continuous fashion because its template strand has 5’ to 3’ directionality, which means the polymerase must work backwards from the replication fork. This causes periodic breaks in the process of creating the lagging strand. The primase and polymerase move in the opposite direction of the fork, so the enzymes must repeatedly stop and start again while the DNA helicase breaks the strands apart. Once the fragments are made, DNA ligase connects them into a single, continuous strand. The entire replication process is considered "semi-discontinuous" since one of the new strands is formed continuously and the other is not.
During the 1960s, Reiji and Tsuneko Okazaki conducted experiments involving DNA replication in the bacterium Escherichia coli. Before this time, it was commonly thought that replication was a continuous process for both strands, but the discoveries involving E. coli led to a new model of replication. The scientists found there was a discontinuous replication process by pulse-labeling DNA and observing changes that pointed to non-contiguous replication. (W)
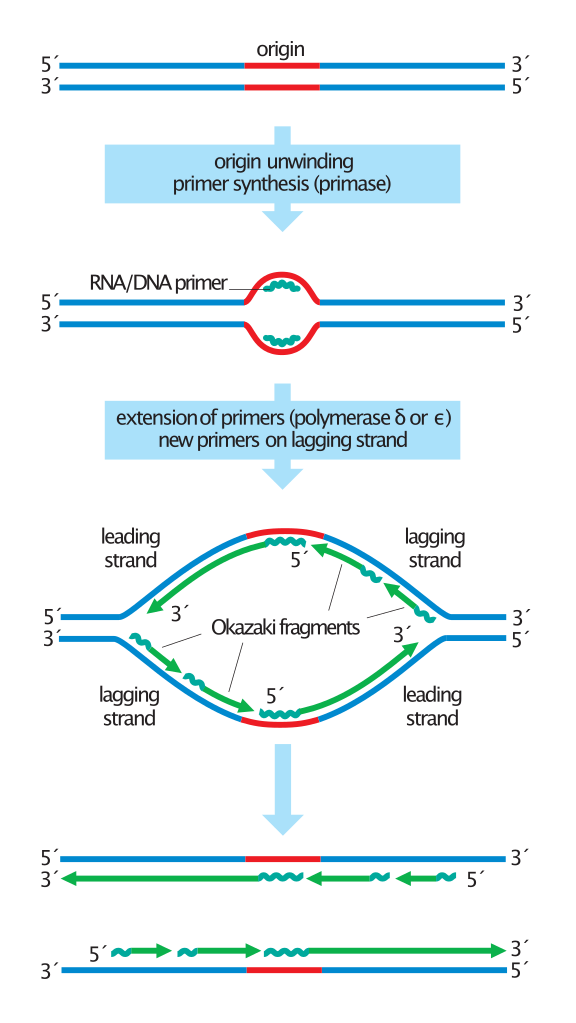
Asymmetry in the synthesis of leading and lagging strands.
Duplication of the DNA begins with origin unwinding, followed by the synthesis of RNA primers (jagged lines) on both DNA strands. DNA polymerase delta or epsilon extends these primers by adding new DNA (green lines) only in a 5' to 3' direction. On the leading strands, this results in the continuous synthesis of long DNA molecules. Lagging strands, in contrast, are synthesized discontinuously: primers are placed on the template every ~200 nucleotides and extended to form short Okazaki fragments. For simplicity, this diagram does not show the replacement of primers with DNA or the synthesis of telomeres at the chromosome ends. |
|

Many enzymes are involved in the DNA replication fork. |
|
Synthesis of Okazaki fragments. |
|
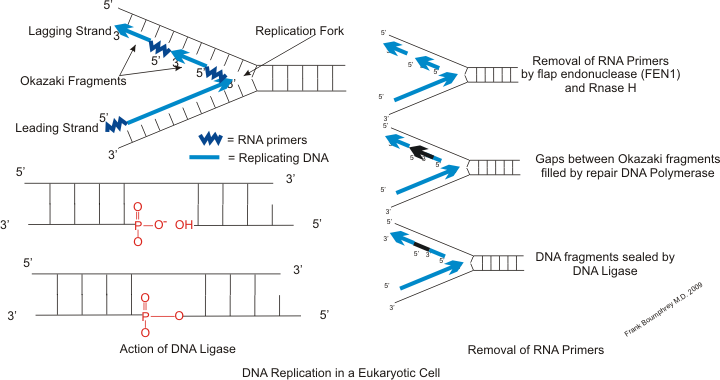
Primase adds RNA primers onto the lagging strand, which allows synthesis of Okazaki fragments from 5' to 3'. However, primase creates RNA primers at a much lower rate than that at which DNA polymerase synthesizes DNA on the leading strand. DNA polymerase on the lagging strand also has to be continually recycled to construct Okazaki fragments following RNA primers. This makes the speed of lagging strand synthesis much lower than that of the leading strand. To solve this, primase acts as a temporary stop signal, briefly halting the progression of the replication fork during DNA replication. This molecular process prevents the leading strand from overtaking the lagging strand. |
|
|
|
DNA replication — Pre-replication complex
A pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of the genome ensures that each daughter cell will carry the same genetic information as the parent cell. Accordingly, formation of the pre-RC is a very important part of the cell cycle. (W)
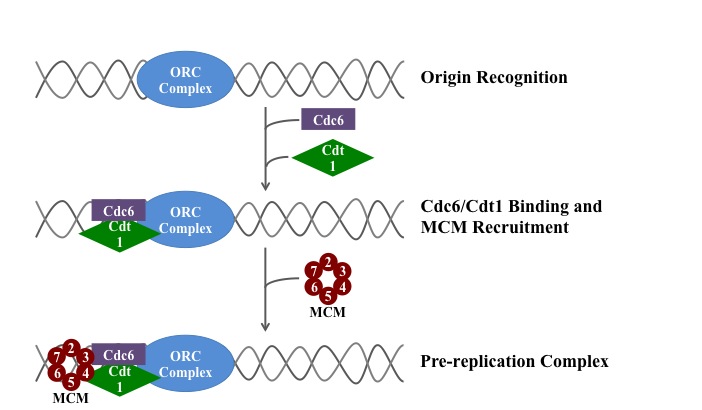
A simplified schematic of the loading of the eukaryotic pre-replication complex. |
|
|
|
DNA replication — Replication through nucleosomes
Eukaryotic DNA must be tightly compacted in order to fit within the confined space of the nucleus. Chromosomes are packaged by wrapping 147 nucleotides around an octamer of histone proteins, forming a nucleosome. The nucleosome octamer includes two copies of each histone H2A, H2B, H3, and H4. Due to the tight association of histone proteins to DNA, eukaryotic cells have proteins that are designed to remodel histones ahead of the replication fork, in order to allow smooth progression of the replisome. There are also proteins involved in reassembling histones behind the replication fork to reestablish the nucleosome conformation.
There are several histone chaperones that are known to be involved in nucleosome assembly after replication. The FACT complex has been found to interact with DNA polymerase α-primase complex, and the subunits of the FACT complex interacted genetically with replication factors. The FACT complex is a heterodimer that does not hydrolyze ATP, but is able to facilitate "loosening" of histones in nucleosomes, but how the FACT complex is able to relieve the tight association of histones for DNA removal remains unanswered. (W)
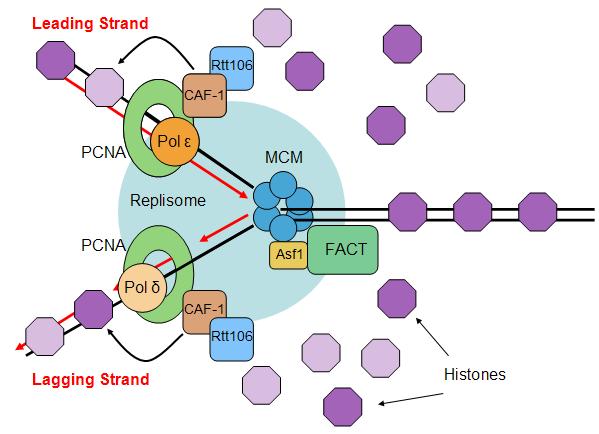
Depiction of replication through histones. Histones are removed from DNA by the FACT complex and Asf1. Histones are reassembled onto newly replicated DNA after the replication fork by CAF-1 and Rtt106. |
|
|
|
DNA replication — Termination
Termination of eukaryotic DNA replication requires different processes depending on whether the chromosomes are circular or linear. Unlike linear molecules, circular chromosomes are able to replicate the entire molecule. However, the two DNA molecules will remain linked together. This issue is handled by decatenation of the two DNA molecules by a type II topoisomerase. Type II topoisomerases are also used to separate linear strands as they are intricately folded into a nucleosome within the cell.
As previously mentioned, linear chromosomes face another issue that is not seen in circular DNA replication. Due to the fact that an RNA primer is required for initiation of DNA synthesis, the lagging strand is at a disadvantage in replicating the entire chromosome. While the leading strand can use a single RNA primer to extend the 5' terminus of the replicating DNA strand, multiple RNA primers are responsible for lagging strand synthesis, creating Okazaki fragments. This leads to an issue due to the fact that DNA polymerase is only able to add to the 3' end of the DNA strand. The 3'-5' action of DNA polymerase along the parent strand leaves a short single-stranded DNA (ssDNA) region at the 3' end of the parent strand when the Okazaki fragments have been repaired. Since replication occurs in opposite directions at opposite ends of parent chromosomes, each strand is a lagging strand at one end. Over time this would result in progressive shortening of both daughter chromosomes. This is known as the end replication problem.
The end replication problem is handled in eukaryotic cells by telomere regions and telomerase. Telomeres extend the 3' end of the parental chromosome beyond the 5' end of the daughter strand. This single-stranded DNA structure can act as an origin of replication that recruits telomerase. Telomerase is a specialized DNA polymerase that consists of multiple protein subunits and an RNA component. The RNA component of telomerase anneals to the single stranded 3' end of the template DNA and contains 1.5 copies of the telomeric sequence. Telomerase contains a protein subunit that is a reverse transcriptase called telomerase reverse transcriptase or TERT. TERT synthesizes DNA until the end of the template telomerase RNA and then disengages. This process can be repeated as many times as needed with the extension of the 3' end of the parental DNA molecule. This 3' addition provides a template for extension of the 5' end of the daughter strand by lagging strand DNA synthesis. Regulation of telomerase activity is handled by telomere-binding proteins. (W)

A depiction of telomerase progressively elongating telomeric DNA. |
|
|
|
DNA replisome
The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
In terms of structure, the replisome is composed of two replicative polymerase complexes, one of which synthesizes the leading strand, while the other synthesizes the lagging strand. The replisome is composed of a number of proteins including helicase, RFC, PCNA, gyrase/topoisomerase, SSB/RPA, primase, DNA polymerase III, RNAse H, and ligase. (W)

A representation of the structures of the replisome during DNA replication. |
|
|
|
DNA sense
Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply sense) if its nucleotide sequence corresponds directly to the sequence of an RNA transcript which is translated or translatable into a sequence of amino acids (provided that any thymine bases in the DNA sequence are replaced with uracil bases in the RNA sequence). The other strand of the double-stranded DNA molecule is referred to as negative-sense (also negative (-) or antisense), and is reverse complementary to both the positive-sense strand and the RNA transcript. It is actually the antisense strand that is used as the template from which RNA polymerases construct the RNA transcript, but the complementary base-pairing by which nucleic acid polymerization occurs means that the sequence of the RNA transcript will look identical to the sense strand, apart from the RNA transcript's use of uracil instead of thymine.
Sometimes the phrases coding strand and template strand are encountered in place of sense and antisense, respectively, and in the context of a double-stranded DNA molecule the usage of these terms is essentially equivalent. However, the coding/sense strand need not always contain a code that is used to make a protein; both protein-coding and non-coding RNAs may be transcribed.
The terms "sense" and "antisense" are relative only to the particular RNA transcript in question, and not to the DNA strand as a whole. In other words, either DNA strand can serve as the sense or antisense strand. Most organisms with sufficiently large genomes make use of both strands, with each strand functioning as the template strand for different RNA transcripts in different places along the same DNA molecule. In some cases, RNA transcripts can be transcribed in both directions (i.e. on either strand) from a common promoter region, or be transcribed from within introns on either strand (see "ambisense" below). (W)
|
|
DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
Knowledge of DNA sequences has become indispensable for basic biological research, and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged.
The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in the sequencing of complete DNA sequences, or genomes, of numerous types and species of life, including the human genome and other complete DNA sequences of many animal, plant, and microbial species.
The first DNA sequences were obtained in the early 1970s by academic researchers using laborious methods based on two-dimensional chromatography. Following the development of fluorescence-based sequencing methods with a DNA sequencer, DNA sequencing has become easier and orders of magnitude faster. (W)
An example of the results of automated chain-termination DNA sequencing. |
|
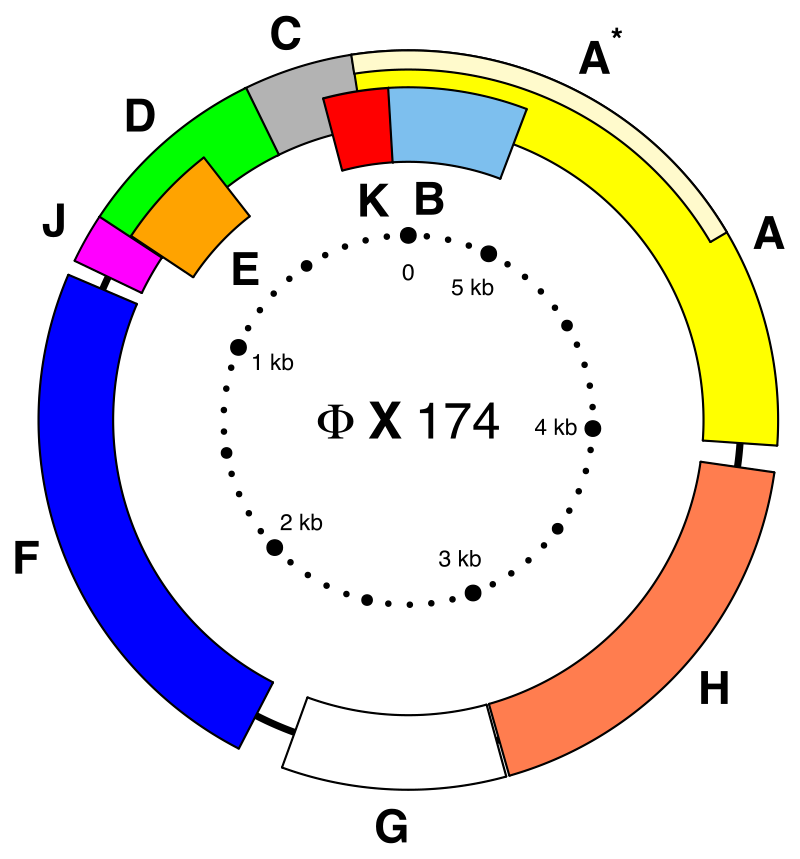
The 5,386 bp genome of bacteriophage φX174. Each coloured block represents a gene.
The map shows the complete circular single-stranded DNA genome (5386 bp) of Enterobacteria phage ΦX174 (accession NC_001422). This DNA genome was the first one ever sequenced (Fred Sanger and colleagues: 1977). This genome contains 11 genes (A, A*, B-H, J, K). Genes B, K, E are overlapping with genes A, C, D. Gene A* is nested within the larger A gene, with internal translation initiation in the same reading frame as gene A. |
|
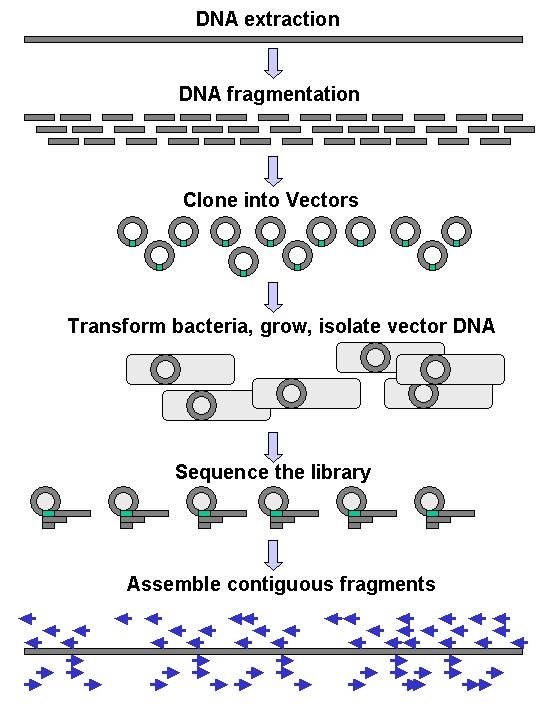
Genomic DNA is fragmented into random pieces and cloned as a bacterial library. DNA from individual bacterial clones is sequenced and the sequence is assembled by using overlapping DNA regions. |
|
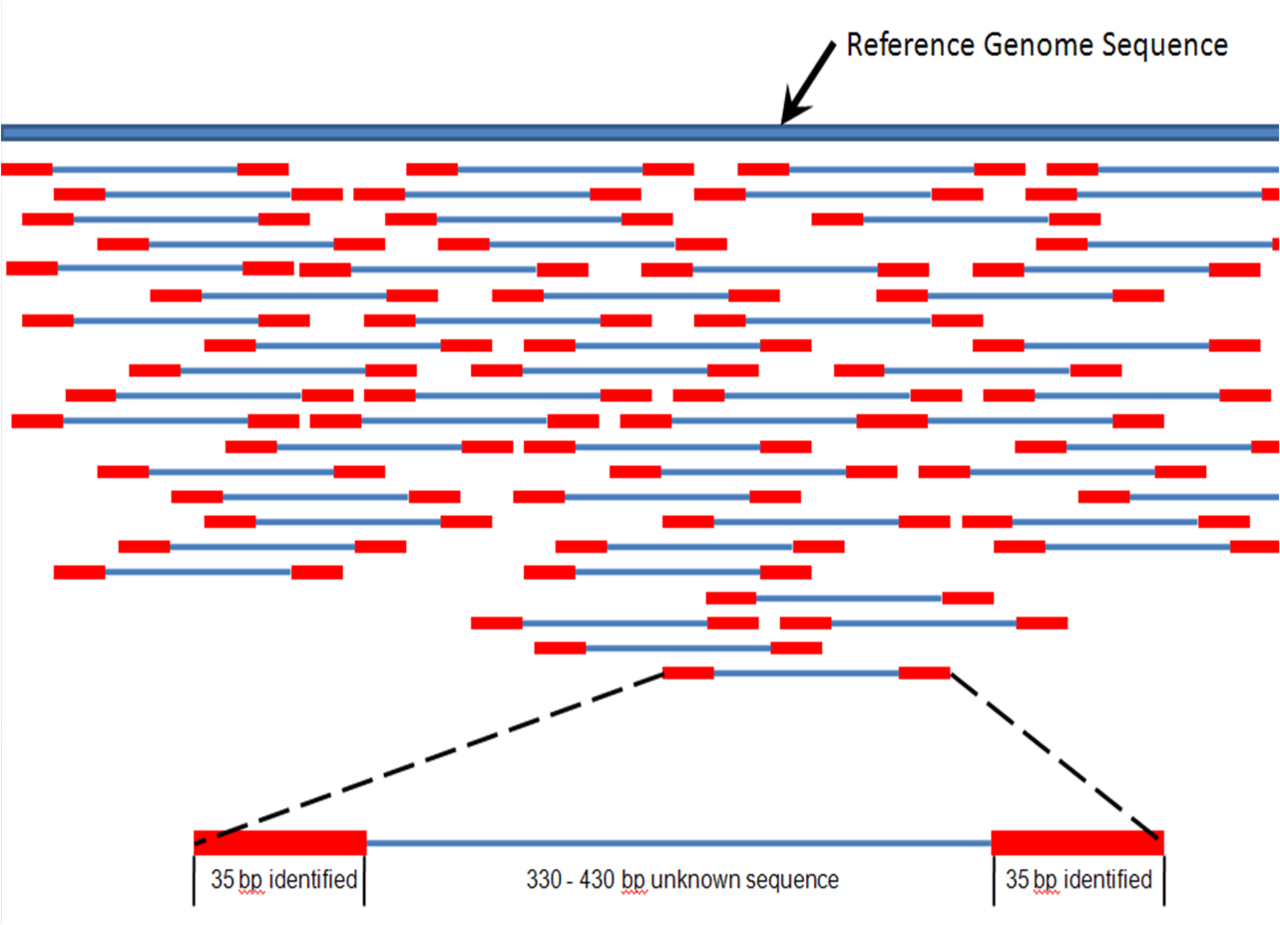
Multiple, fragmented sequence reads must be assembled together on the basis of their overlapping areas. |
|

An Illumina HiSeq 2500 sequencer (photo taken at the Max Planck Genomzentrum Cologne, Germany). |
|

A BGI MGISEQ-2000RS sequencer. |
|
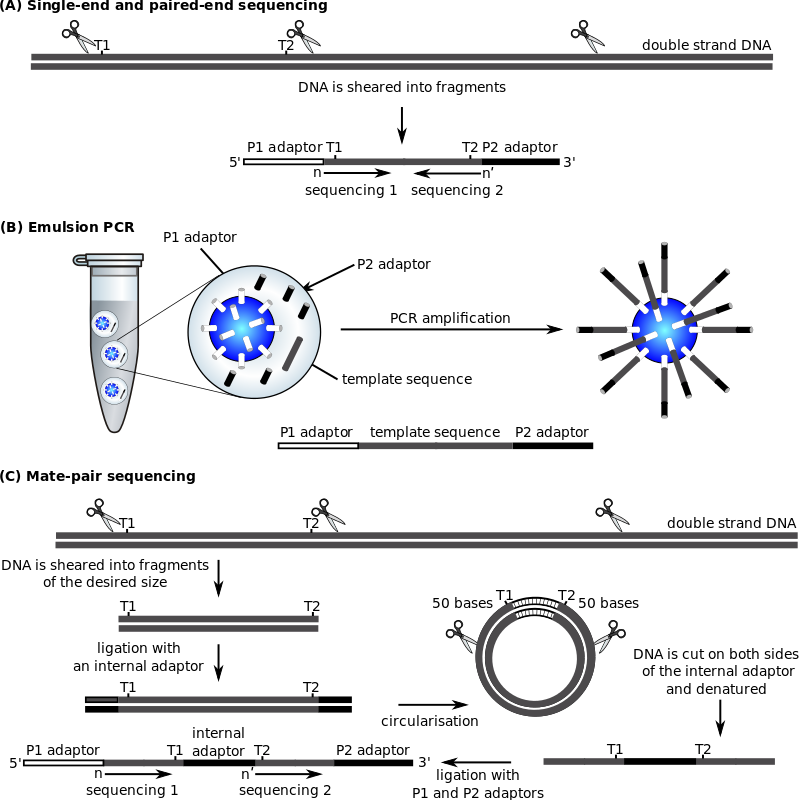
Library preparation for the SOLiD platform. |
|
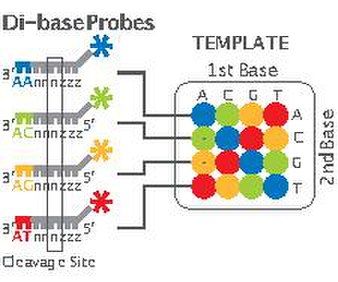
Two-base encoding scheme. In two-base encoding, each unique pair of bases on the 3' end of the probe is assigned one out of four possible colors. For example, "AA" is assigned to blue, "AC" is assigned to green, and so on for all 16 unique pairs. During sequencing, each base in the template is sequenced twice, and the resulting data are decoded according to this scheme. |
|
|
|
DNA sequencing theory
DNA sequencing theory is the broad body of work that attempts to lay analytical foundations for determining the order of specific nucleotides in a sequence of DNA, otherwise known as DNA sequencing. The practical aspects revolve around designing and optimizing sequencing projects (known as "strategic genomics"), predicting project performance, troubleshooting experimental results, characterizing factors such as sequence bias and the effects of software processing algorithms, and comparing various sequencing methods to one another. In this sense, it could be considered a branch of systems engineering or operations research. The permanent archive of work is primarily mathematical, although numerical calculations are often conducted for particular problems too. DNA sequencing theory addresses physical processes related to sequencing DNA and should not be confused with theories of analyzing resultant DNA sequences, e.g. sequence alignment. Publications sometimes do not make a careful distinction, but the latter are primarily concerned with algorithmic issues. Sequencing theory is based on elements of mathematics, biology, and systems engineering, so it is highly interdisciplinary. The subject may be studied within the context of computational biology. (W) |
|
DNA virus
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: Duplodnaviria and Varidnaviria, and ssDNA viruses are almost exclusively assigned to the realm Monodnaviria, which also includes dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Viruses that have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase are separately considered reverse transcribing viruses and are assigned to the kingdom Pararnavirae in the realm Riboviria.
DNA viruses are ubiquitous worldwide, especially in marine environments where they form an important part of marine ecosystems, and infect both prokaryotes and eukaryotes. They appear to have multiple origins, as viruses in Monodnaviria appear to have emerged from archaeal and bacterial plasmids on multiple occasions, though the origins of Duplodnaviria and Varidnaviria are less clear. Prominent disease-causing DNA viruses include herpesviruses, papillomaviruses, and poxviruse. (W)
Illustrated sample of Duplodnaviria virions.
Computer-generated illustration showing a sample of virion morphological diversity in the realm Duplodnaviria. From left to right: Siphoviridae (Caudovirales), Myoviridae (Caudovirales), Podoviridae (Caudovirales), Herpesviridae (Herpesvirales). . |
|
A ribbon diagram of the MCP of Pseudoalteromonas virus PM2, with the two jelly roll folds colored in red and blue.
The major capsid protein P2 from the bacteriophage PM2, illustrating the double jelly roll fold found in the major capsid proteins of double-stranded DNA viruses of the PRD1-adenovirus lineage. The two distinct jelly roll domains are colored red and blue, with intervening loops and helices colored orange. The topology and connectivity in each jelly roll is identical to that illustrated in File:4oq9 chainA jellyroll.png. The trimeric assembly is illustrated in File:2w0c_trimer.png and the full capsid in File:2w0c_capsid.png. Rendered from chain H of PDB ID 2W0C: Abrescia NG, Grimes JM, Kivelä HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JK, Bamford DH, Stuart DI. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Molecular cell. 2008 Sep 5;31(5):749-61. DOI 10.1016/j.molcel.2008.06.026. |
|
|
|
DNase I hypersensitive site
In genetics, DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the DNase I enzyme. In these specific regions of the genome, chromatin has lost its condensed structure, exposing the DNA and making it accessible. This raises the availability of DNA to degradation by enzymes, such as DNase I. These accessible chromatin zones are functionally related to transcriptional activity, since this remodeled state is necessary for the binding of proteins such as transcription factors.
Since the discovery of DHSs 30 years ago, they have been used as markers of regulatory DNA regions. These regions have been shown to map many types of cis-regulatory elements including promoters, enhancers, insulators, silencers and locus control regions. A high-throughput measure of these regions is available through DNase-Seq. (W)
|
|
double-stranded RNA viruses
Double-stranded RNA viruses (dsRNA viruses) are a polyphyletic group of viruses that have double-stranded genomes made of ribonucleic acid. The double-stranded genome is used to transcribe a positive-strand RNA by the viral RNA-dependent RNA polymerase (RdRp). The positive-strand RNA may be used as messenger RNA (mRNA) which can be translated into viral proteins by the host cell's ribosomes. The positive-strand RNA can also be replicated by the RdRp to create a new double-stranded viral genome.
Double-stranded RNA viruses are classified in two separate phyla Duplornaviricota and Pisuviricota (specifically class Duplopiviricetes), which are in the kingdom Orthornavirae and realm Riboviria. The two groups do not share a common dsRNA virus ancestor. Double-stranded RNA viruses evolved two separate times from positive-strand RNA viruses. In the Baltimore classification system, dsRNA viruses belong to Group III.
Virus group members vary widely in host range (animals, plants, fungi, and bacteria), genome segment number (one to twelve), and virion organization (T-number, capsid layers, or turrets). Double-stranded RNA viruses include the rotaviruses, known globally as a common cause of gastroenteritis in young children, and bluetongue virus, an economically significant pathogen of cattle and sheep. The family Reoviridae is the largest and most diverse dsRNA virus family in terms of host range. (W)
Electron micrograph of rotaviruses. The bar = 100 nm.
Note the wheel-like appearance of some of the rotavirus particles. The observance of such particles gave the virus its name ('rota' being the Latin word meaning wheel). Bar = 100 nanometers. Source: Cell culture. Method: Negative-stain Transmission Electron Microscopy. |
|
|
|
duplex sequencing
Duplex sequencing is a library preparation and analysis method for next-generation sequencing (NGS) platforms that employs random tagging of double stranded DNA to detect mutations with higher accuracy and lower error rate. This method uses degenerate molecular tags in addition to sequencing adapters to recognize reads originating from each strand of DNA. The generated sequencing reads then will be analyzed using two methods: single strand consensus sequences (SSCSs) and Duplex consensus sequences (DCSs) assembly. Duplex sequencing theoretically can detect mutations with frequencies as low as 5 x 10−8 that is more than 10,000 fold higher in accuracy compared to the conventional next-generation sequencing methods (W)
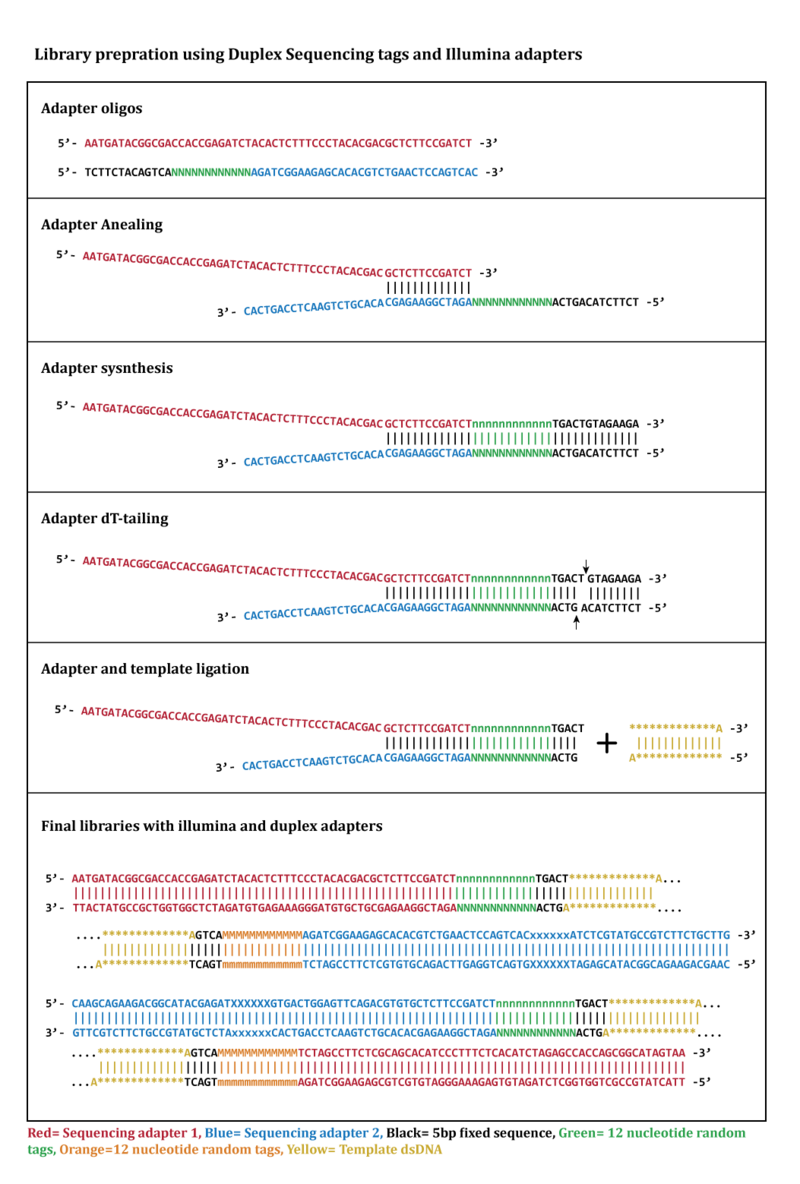
Figure 1) Duplex sequencing library preparation workflow: Two adapter oligos go through several steps (Annealing, Synthesis, dT-tailing) to generate double stranded unique tags with 3'-dT-overhangs. Then the duplex tag adapters ligate to the double stranded DNA templates. Finally Illumina sequencing adapters are inserted into the tagged-DNA fragments and form the final libraries containing DS adapters, Illumina sequencing adapters and template DNA. |
|
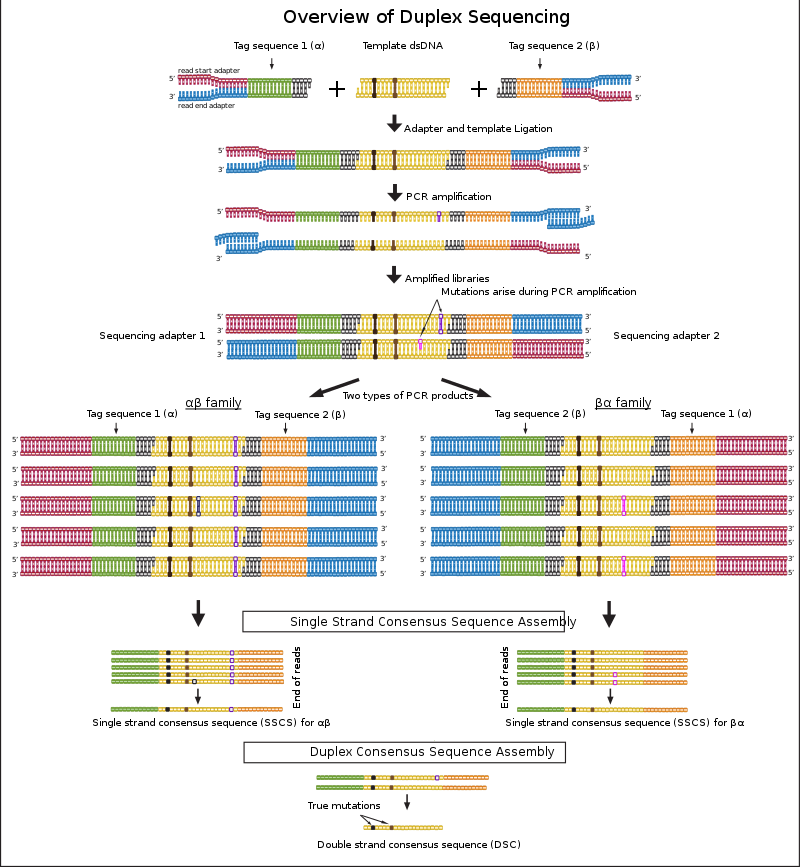
Figure 2) Duplex sequencing overview: Duplex tagged libraries containing sequencing adapters are amplified and result in two types of products each originates from a single strand of DNA. After sequencing the PCR products, the generated reads divide into tag families based on the genomic position, duplex tags and the neighboring sequencing adapter. Please note that sequence tag α is the reverse complement of sequence tag β and vice versa. |
|
|
|